A recently published paper (Kornai et al., 2024, Systematic Biology 73: 1015–1037, linked below) represents a significant improvement in species delimitation methods under the multispecies coalescent model, specifically by incorporating gene flow/migration into the model. This will greatly help distinguish continuous geographic variation from actual species boundaries.
A recent trend has been to use basic MSC models to split geographic variation in widespread groups into many local “species,” despite continuous gene flow among the local populations. Many MSC models that make simplifying assumptions will split almost any population samples into different groups, which has led to some extreme cases of over-splitting in recent years, especially in herps and fishes, but in other groups as well. We explored this problem in Chambers and Hillis, 2020: Syst. Biol. 69:184–193, The Multispecies Coalescent Over-Splits Species in the Case of Geographically Widespread Taxa.
In response, the authors of this new method propose a range of results, depending on assumptions that are made. Somewhat confusingly, their “merge” algorithm tends to be more likely to split species, and the “split” algorithm tends to be more likely to merge them. But exploring a range of options is a big improvement, as are assumptions that incorporate gene flow. When the results of the algorithms differ, users presumably need to rely on additional data (such as the analysis of contact zones between the putative taxa) to decide on their preferred taxonomic arrangement.
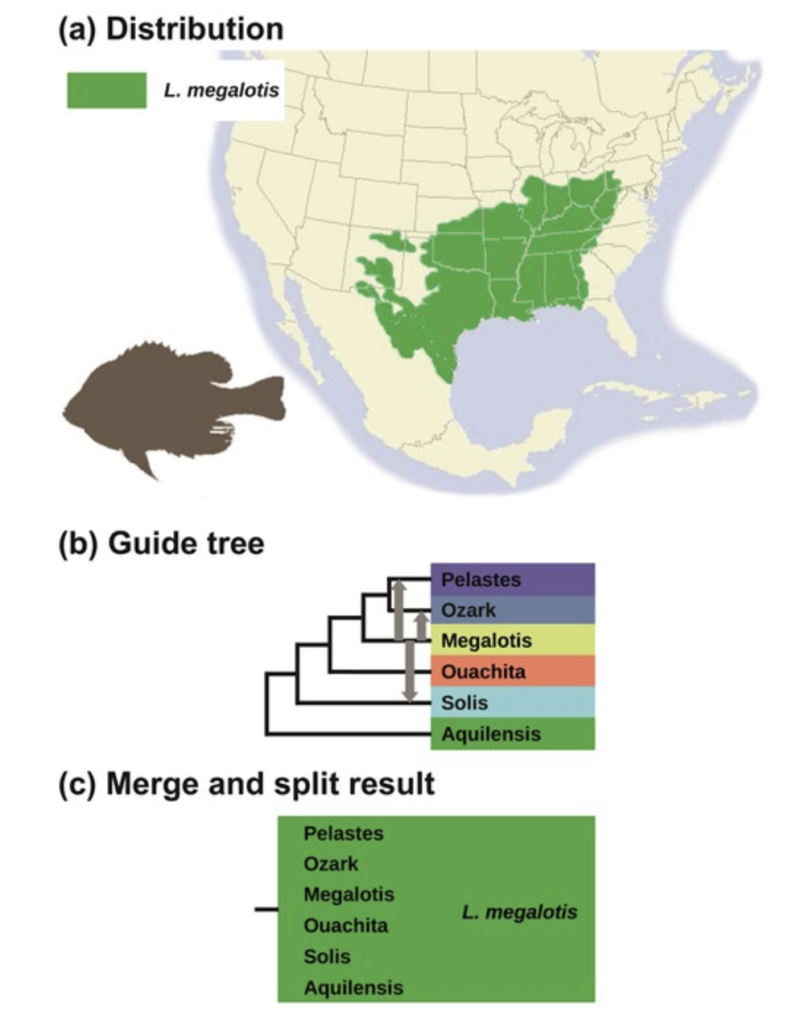
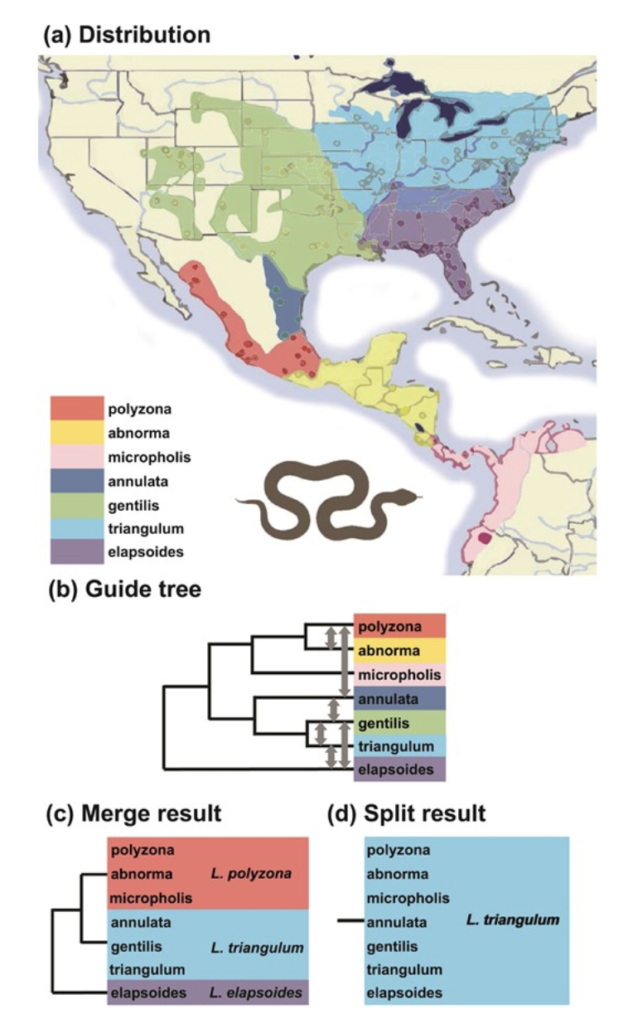
The authors applied their method to several recent controversial taxonomic splits of geographically variable, widespread species. These included the American Milksnakes (Lampropeltis triangulum complex), and the Longear Sunfish (Lepomis megalotis), both of which have been proposed to be split into many species by some recent authors.
In the case of the Milksnakes, the new method proposes either three species (as proposed by Chambers and Hillis, 2020), or a single species (as proposed by Williams, 1988, in his monograph on the complex). It provides no support for the recognition of seven species (as proposed by Ruane et al., 2014: Syst. Biol. 63:231–250). So, how does one decide between the one and three species conclusions? As we noted in Chambers at al. (2023: Syst. Biol. 72:357–371), the contact zones of the putative taxa should be examined and analyzed. In this case, there is clear evidence of reproductive isolation (and sympatry of the species) at the contact zone between L. triangulum and L. elapsoides on one hand, and between L. triangulum and L. polyzona on the other. Other contact zones between subspecies show a gradual continuum of intergradation and gene exchange. Thus, the three-species model has strong support, and the one-species and seven-species models can be rejected.
In the case of the Longear Sunfish, the results from this new method are unambiguous. There is no support for dividing Lepomis megalotis into multiple regional species, using either the conservative or the liberal algorithms.