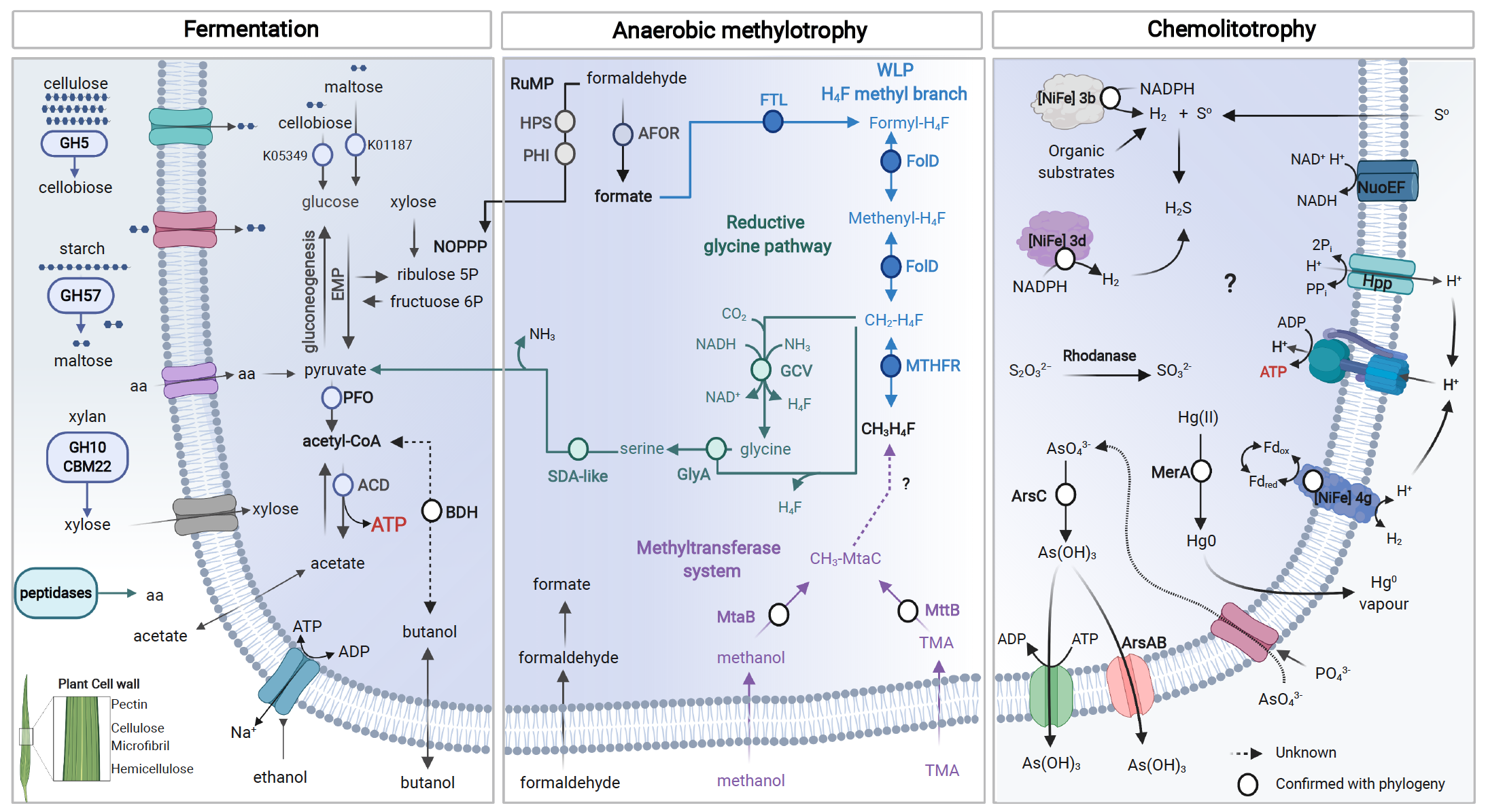
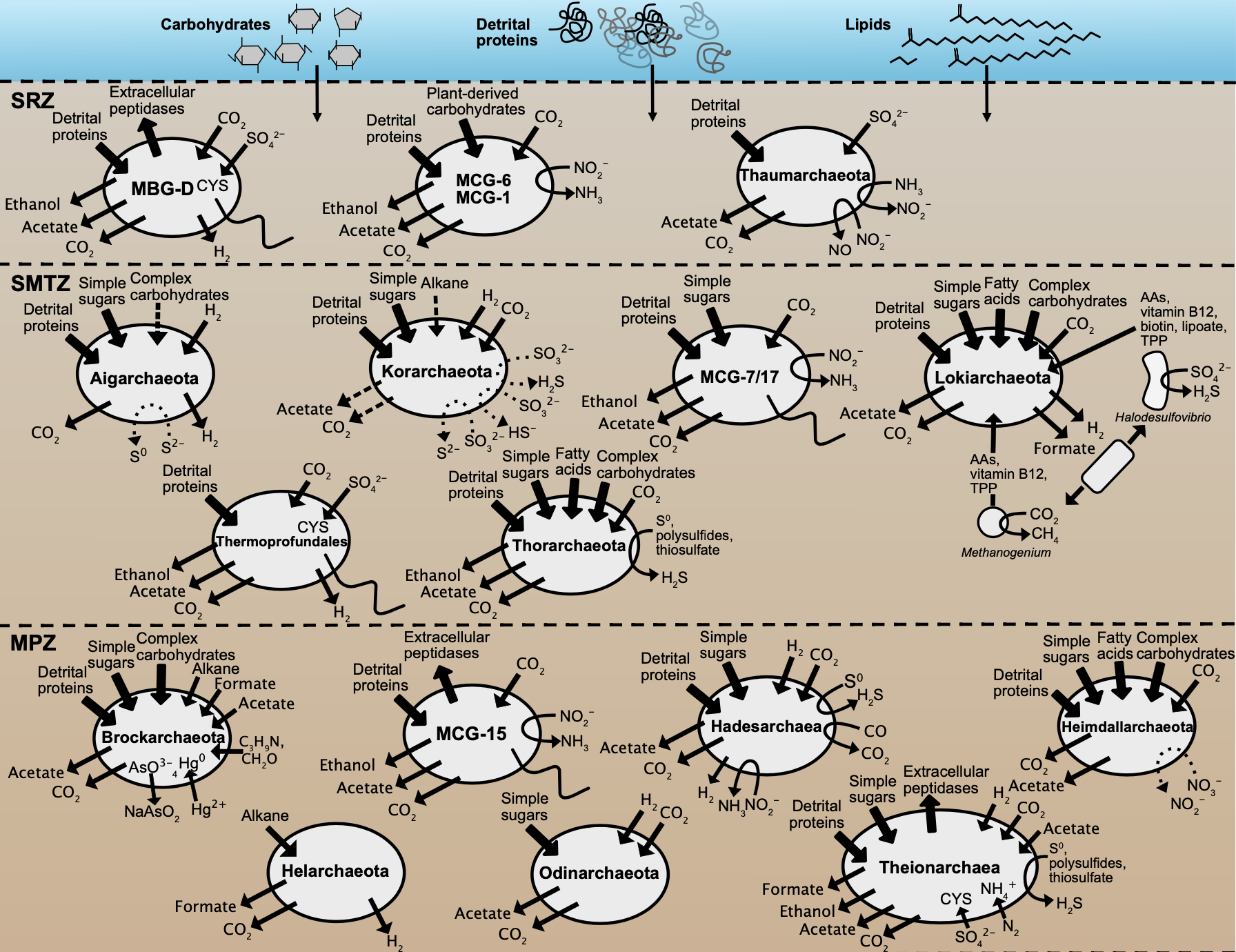
Summary
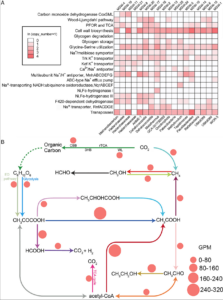
Reduced substrates produced by the serpentinization reaction under hydration of olivine may have fuelled biological processes on early Earth. To understand the adaptive strategies and carbon metabolism of the microbes in the serpentinizing ecosystems, we reconstructed 18 draft genomes representing dominant species of Omnitrophicaeota, Gammaproteobacteria and Methanobacteria from the Manleluag serpentinizing spring in Zambales, Philippines (hyperalkaline and rich in methane and hydrogen). Phylogenomics revealed that two genomes were affiliated with a candidate phylum NPL-UPA2 and the references of all our genomes were derived from ground waters, hot springs and the deep biosphere. C1 metabolism appears to be widespread as most of the genomes code for methanogenesis, CO oxidation and CO2 fixation. However, likely due to the low CO2concentration and election acceptors, the biomass in the spring was extremely low (<103 cell/ml). Various Na+ and K+ transporters and Na+-driving ATPases appear to be encoded by these genomes, suggesting that nutrient acquisition, bioenergetics and normal cytoplasmic pH were dependent on Na+ and K+ pumps. Our results advance our understanding of the metabolic potentials and bioenergetics of serpentinizing springs and provide a framework of the ecology of early Earth.