“…evolution is the control of development by ecology”
Leigh Van Valen. 1973. Science.
integrative approaches to convergence and diversification
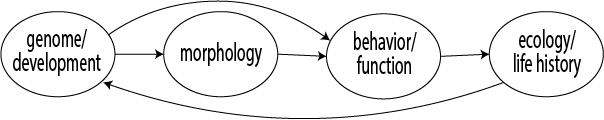
Evolution often repeats itself. This is evident in many examples, including morphological convergence of pitcher traps in carnivorous plants, diet and jaw specializations in cichlid fishes in east African lakes, and ecological convergence in marsupial and placental mammals. We know that genes and molecular pathways can be preserved for hundreds of millions of years. These conserved molecular and developmental tools can direct the evolution of similar traits in response to similar ecological conditions. However, in other cases, even homologous phenotypes evolved from a common ancestor can arise via diverse developmental and genomic processes. Thus, capturing the origin and evolution of biodiversity requires an integrative lens spanning organismal and evolutionary levels. In our lab, we use diverse vertebrate systems to answer three overarching questions about the development and evolution of variation in form and function.
How is phenotypic diversity linked to development, genomic, & ecological variation?

Frogs have diverse reproductive adaptations. Apart from the ancestral aquatic embryos, species have independently evolved behaviors (e.g., nest building and parental care) and terrestrial reproductive modes (e.g., direct development or brooding of embryos in skin pouches). We identify the genomic mechanisms underlying the differentiation of embryogenesis in the “terrestrially” reproducing species. In collaboration with Dr. Andrés Romero-Carvajal, we further ask whether independent transitions to terrestrial reproduction are associated with convergent embryonic modifications.

Maternal effects link offspring development to current environmental conditions and thus, provide a potential mechanism for the development of locally appropriate phenotypic plasticity. In novel environments, mothers can increase the survival of offspring through locally- adaptive and sex-specific maternal responses. Coupling ‘omics approaches like proteomic and metabolomic methods that quantify protein and hormonal resources extracted from avian yolks with transcriptomic analysis of early embryonic tissue, enabling the discovery of how maternal strategies and offspring responses vary across environments or in a sex-specific way.
How commonly do shared genomic pathways lead to phenotypic convergence?
Despite the remarkable diversity of life forms on earth, evolutionary biologists have discovered numerous instances where even distantly related species share astonishing similarities in how they behave, look, and function. Given the importance of happenstance in evolution (e.g., random mutations, genetic drift, environmental stochasticity), it is often assumed that the mechanisms underlying such convergent phenotypes are distinct. Nevertheless, recent discoveries that the same pathways can underlie convergently evolved phenotypes have reinvigorated questions about the predictability of evolution and whether broadly conserved genomic mechanisms facilitate phenotypic convergence. Capitalizing on diverse cases of convergent evolution including monogamous mating systems, mutualistic cleaning behavior, social dominance, and embryonic constraints, we integrate comparative ‘omics, comparative developmental biology, and computational biology to identify mechanisms of convergent evolution.
Why do some traits vary more than others?
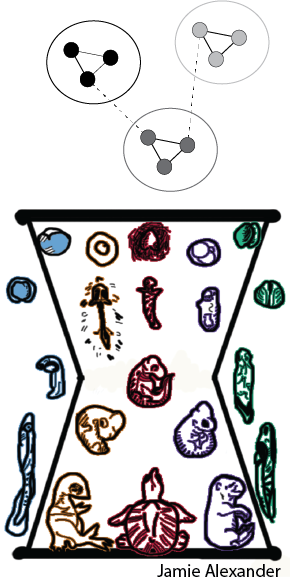
Complex traits consist of multiple components, each with potentially independent evolutionary histories that can vary within and across species. Why do some traits seem more evolutionarily flexible than others? We posit that semi-independence of developmental and/or genomic underpinnings may enable both within-species variability and between-species flexibility of complex traits. Our lab is particularly interested in whether the modularity of gene regulatory networks (GRNs) underlies evolutionary flexibility observed in complex traits like embryogenesis, behavioral systems, and morphological characters.
