Est. in 2008 at
The University of Texas at Austin
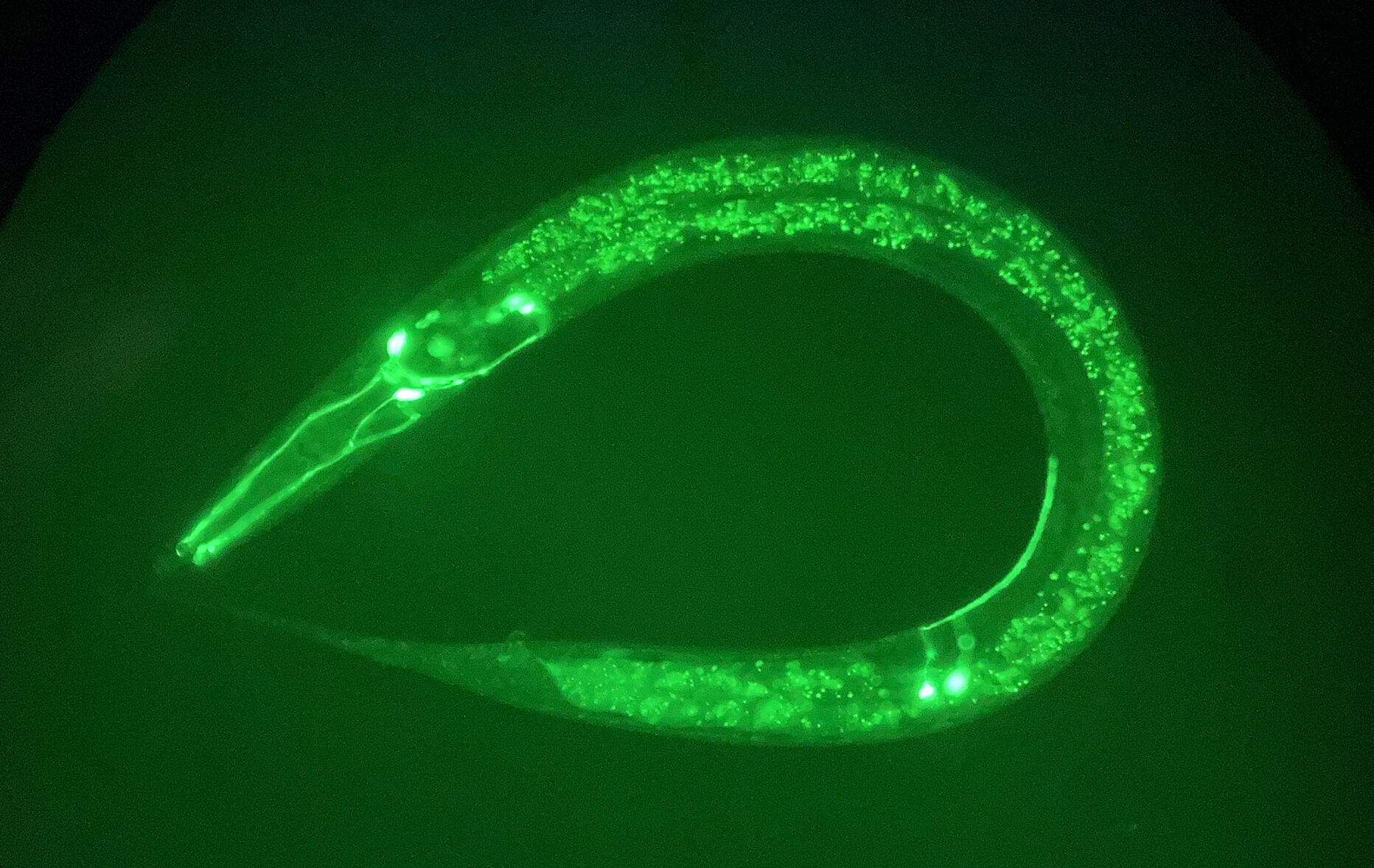
Using the simple but powerful model nematode, C. elegans, to investigate genetic mechanisms that govern behaviors and contribute to neurological disorders.
“BZ555” Photo credit- Lisa Wang
Our Research
Which genes cause problems in Down syndrome? Dr. Pierce became interested in Down syndrome (DS) when his son was born with this condition. To check if C. elegans could be used to study DS, our lab compared genes on the 21st chromosome (Hsa21) to worm genes. We found that excluding keratin genes, C. elegans has equivalents for at least half of the remaining genes, including 48 that are highly conserved (Nordquist et all., 2018). To determine the role of these genes, we systematically knocked out or down each one in worm. Of the identified genes, we showed that 15 are needed for viability and that 10 are required for the normal function of the nervous system. We found that two of the unstudied genes, mtq-2 and pdxk-1, were enriched in the nervous system and required for proper synaptic transmission. We are beginning to discover the function of these unstudied molecules. In parallel, we are testing which Hsa21 genes cause phenotypes by overexpressing them in worm. In the process, we have uncovered general principles that explain which genes throughout our genome cause medical problems when overexpressed. Our goal is to identify genes and pathways to target to improve quality of life for people with DS.


Can we stop neurodegeneration in Alzheimer’s disease? Alzheimer’s disease (AD) is inevitable in DS due to an extra copy of the Amyloid Precursor Protein (APP) gene. We have engineered C. elegans to mimic the key features of neurodegeneration observed in human AD by overexpressing a single copy of APP (Yi et al., 2017). We can now test with unprecedented speed, whether drugs prevent degeneration by simply counting fluorescent neurons through the worm’s transparent body (Modal et al., 2018). While mouse studies of AD typically take 2 years, the compact lifespan of C. elegans affords studies as short as 1 week. With our new powerful model of AD, we are learning why certain types of neurons are vulnerable in AD, how they die, and how APOE variants and TAU influence vulnerability to AD (Sae-Lee et al., 2020; Cardona et al., 2025). During these studies, we discovered a surprising role for EGL-1 – the trigger for apoptotic cell-death in all organisms. Rather than killing, we found that it unexpectedly protects neurons by triggering ejection of excess proteins and organelles in large membrane-bound blebs called exophers (Wu et al., 2025).
How does alcohol affect our nervous system? How we get drunk remains unclear at the molecular level. By using C. elegans, we can rapidly identify molecular mechanisms responsible for behavioral responses to alcohol. For instance, we completed unbiased forward-genetic and GWAS screens to identify the BK potassium channel as the single most important single molecule required for intoxication in C. elegans (Davies et al., 2003). We also discovered how alcohol activates the human BK potassium channel (Davis et al., 2014), and developed drugs to interfere with this response that relieve withdrawal in rodents (Scott et al,. 2018; Scott et al., 2018). Further, we tested the worm’s alcohol-insensitive mutation in the BK channel of mouse. This knock-in mouse is selective resistant to alcohol but not other sedative drugs. This provides strong evidence that the BK channel is a major target for alcohol action in mammals.


Which genes cause autism? How do mutations in >100 genes disrupt nervous system function to cause atypical social and sensory behaviors observed in ASD? Our lab has found that the severity and number of mutations in these genes causally decrease the degree of social behavior of C. elegans. Conversely, we found that mutation of genes associated with hypersocial condition, Williams syndrome, increases social behavior in worm. Thus, unexpected conservation of social behavior allows us to model ASD in worm. We are using this new paradigm to study how ASD genes interact and, in collaboration, succeeded in screening drugs that reduce phenotypes in patient models of ASD.
