Baker lab leading a new project to understand the origin of eukaryotes funded by the Simons and Moore Foundations
New Initiative Aims to Reveal the Origins of Complex Life
A new research initiative will shed light on how the origin of complex life evolved through symbiosis. The project will be the latest from the Brett Baker laboratory at The University of Texas at Austin’s Marine Science Institute, which made recent discoveries of new organisms called Asgard archaea, named after Norse gods, and their metabolisms.
The Asgards are commonly found at the bottom of the ocean near hydrothermal vents, and these discoveries brought scientists closer to understanding how more complex life, known as eukaryotes, came to be. The Moore–Simons Project on the Origin of the Eukaryotic Cell has awarded a grant to Baker and his colleagues (Dr. Thijs Ettema Wageningen at University in the Netherlands, Dr. Mark Ellisman at University of California San Diego, and Dr. Roland Hatzenpichler at Montana State University) to prove or disprove ideas about the origin of complex life.
There is currently a fundamental gap in the understanding of the origin of eukaryotes (including plants and animals). The leading theory is that eukaryotes, with their complex cells full of organelles encased in a cellular membrane, evolved when one simple organism absorbed another, its bacterial symbiont. The recently discovered Asgard archaea are the closest relatives of eukaryotes known and are a descendant of the host of this relationship that led to the first eukaryotic cells. Studies of these Asgard microbes are revolutionizing our understanding of the origin of eukaryotic life.
There are several competing hypotheses on the biological and physiological interactions that led to the symbiotic origin of eukaryotes, and the newly funded research will advance our understanding of this relationship. Scientists know that the evolution of mitochondria material was a pivotal point in eukaryotic evolution because these organelles have their own DNA and are important powerhouses of the eukaryotic cell, creating most of the chemical energy. Recent evidence collected by this team indicate that this absorption may have first occurred in an Asgard host microbe when it took over a bacteria – that has the genes and proteins to produce energy.
The new initiative, supported with nearly $2 million from the Moore-Simons Project on the Origin of the Eukaryotic Cell, will allow Baker and colleagues to determine which Asgards are most closely related to eukaryotes and further explore their physiological interactions, and cellular structure.
“Our research will transform our understanding of the microbial interactions that led to the formation of eukaryotic life on the planet,” says Baker.
New review in press at Annual Review in Marine Science
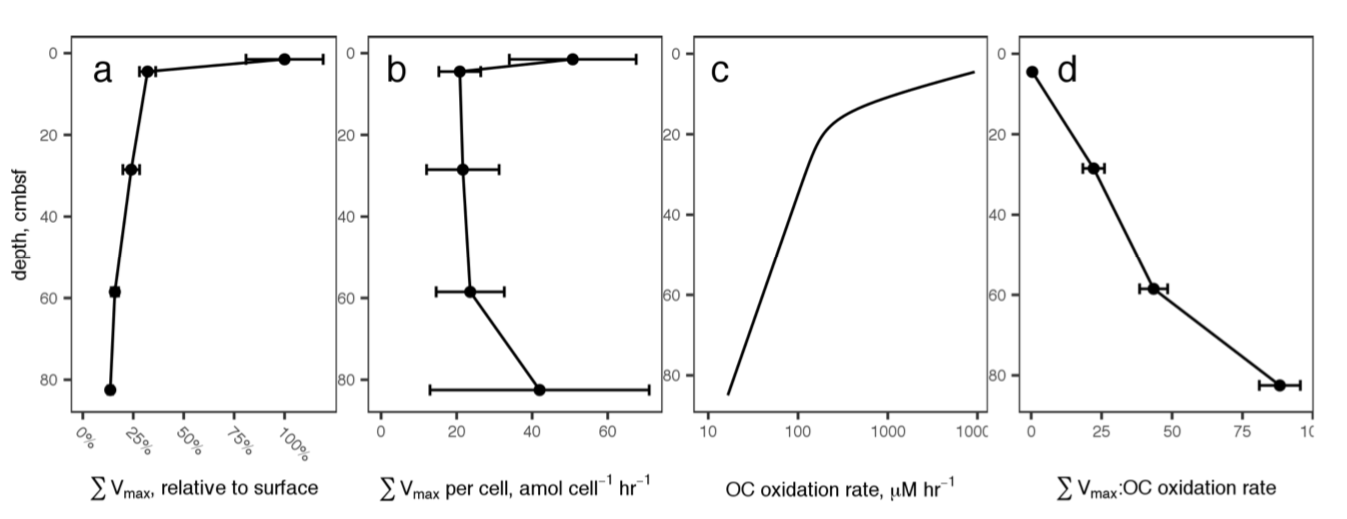
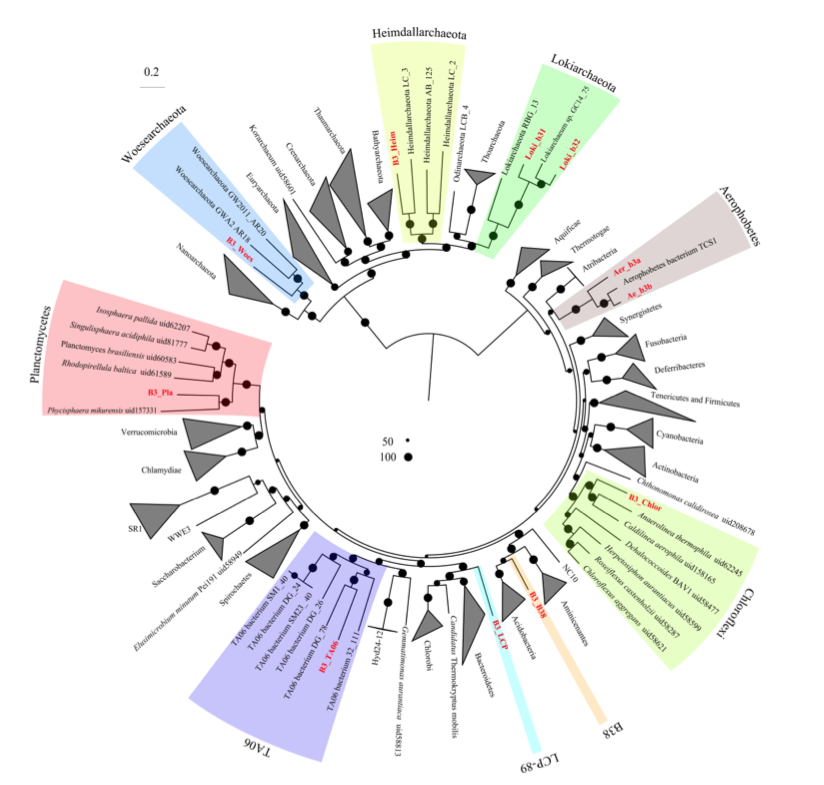
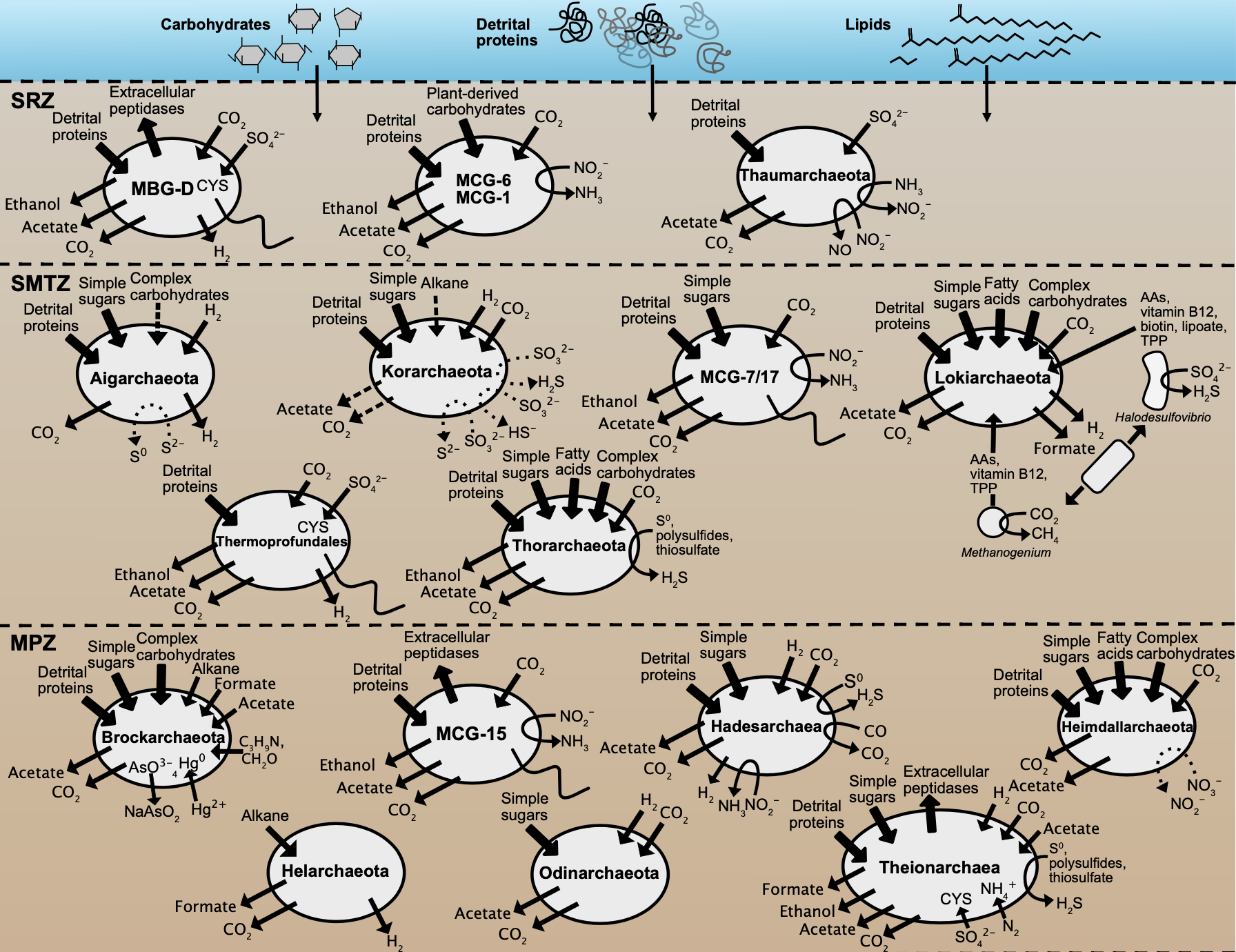
New microbial biodiversity in marine sediments
Microbes in marine sediments represent a large portion of the biosphere, and resolving their ecology is crucial for understanding global ocean processes. Single-gene diversity surveys have revealed several uncultured lineages that are widespread in ocean sediments and whose ecological roles are unknown, and advancements in the computational analysis of increasingly large genomic data sets have made it possible to reconstruct individual genomes from complex microbial communities. Using these metagenomic approaches to characterize sediments is transforming our view of microbial communities on the ocean floor and the biodiversity of the planet. In recent years, marine sediments have been a prominent source of new lineages in the tree of life. The incorporation of these lineages into existing phylogenies has revealed that many belong to distinct phyla, including archaeal phyla that are advancing our understanding of the origins of cellular complexity and eukaryotes. Detailed comparisons of the metabolic potentials of these new lineages have made it clear that uncultured bacteria and archaea are capable of mediating key previously undescribed steps in carbon and nutrient cycling.
Big thanks to the Simons Foundation for this generous funding to our group!
A new review about archaea published in Nature Microbiology
Diversity, ecology, and evolution of Archaea
In this review we explored all the new ecological roles, and new evolutionary insights that have been described in archaea and have generated an updated archaeal tree of life.

Congrats Ian Rambo on his publication about HAB microbiomes!
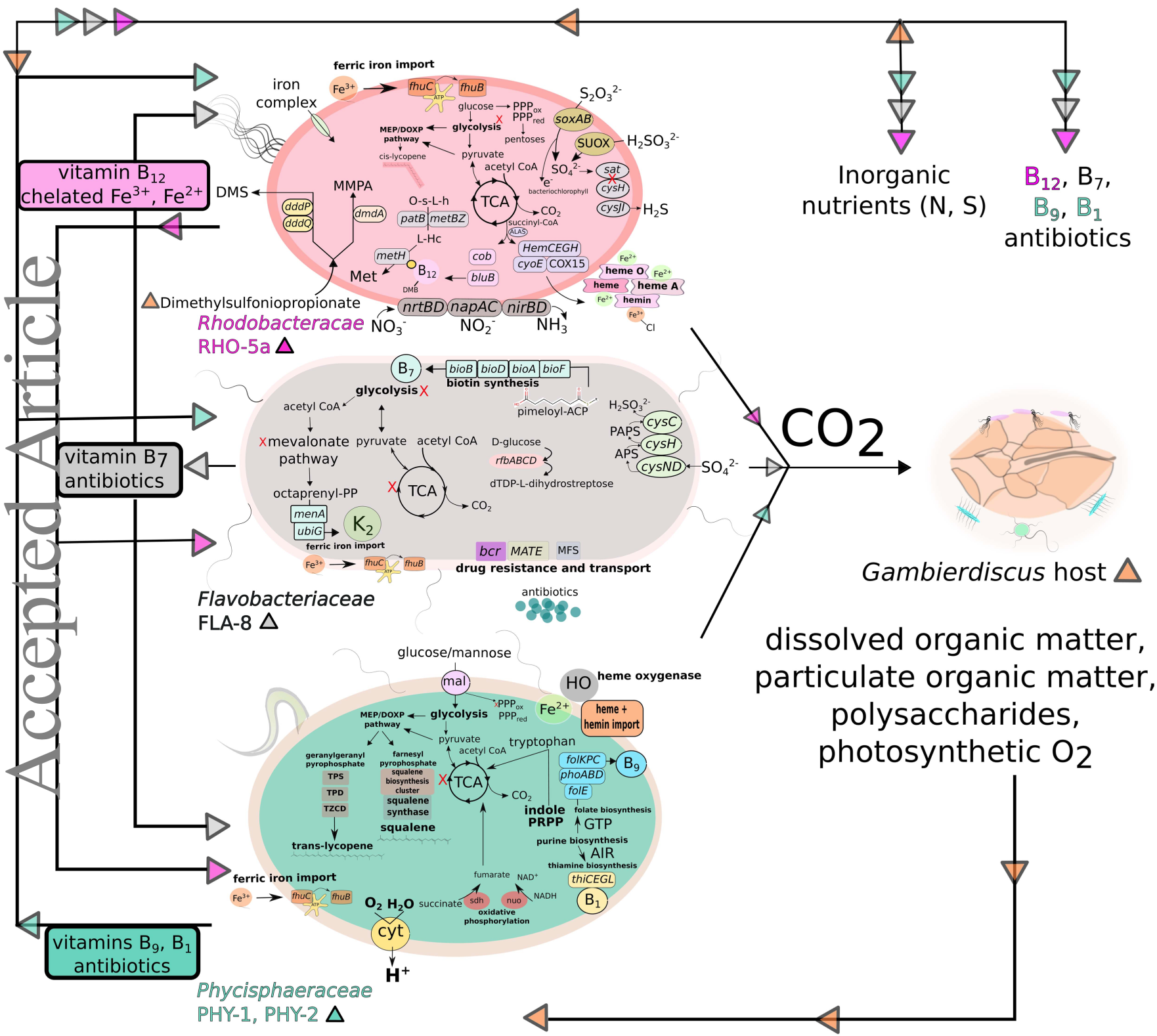
Metabolic relationships of uncultured bacteria associated with the microalgae Gambierdiscus
Summary
Microbial communities inhabit algae cell surfaces and produce a variety of compounds that can impact the fitness of the host. These interactions have been studied via culturing, single‐gene diversity, and metagenomic read survey methods that are limited by culturing biases and fragmented genetic characterizations. Higher‐resolution frameworks are needed to resolve the physiological interactions within these algal‐bacterial communities. Here, we infer the encoded metabolic capabilities of four uncultured bacterial genomes (reconstructed using metagenomic assembly and binning) associated with the marine dinoflagellates Gambierdiscus carolinianus and G. caribaeus. Phylogenetic analyses revealed that two of the genomes belong to the commonly algae‐associated families Rhodobacteraceae and Flavobacteriaceae. The other two genomes belong to the Phycisphaeraceae and include the first algae‐associated representative within the uncultured SM1A02 group. Analyses of all four genomes suggest these bacteria are facultative aerobes, with some capable of metabolizing phytoplanktonic organosulfur compounds including dimethylsulfoniopropionate and sulfated polysaccharides. These communities may biosynthesize compounds beneficial to both the algal host and other bacteria, including iron chelators, B vitamins, methionine, lycopene, squalene, and polyketides. These findings have implications for marine carbon and nutrient cycling and provide a greater depth of understanding regarding the genetic potential for complex physiological interactions between microalgae and their associated bacteria.