Bacterial ecology and evolution converge on seasonal and decadal scales
Robin R. Rohwer1*, Mark Kirkpatrick1, Sarahi L. Garcia2,3, Matthew Kellom4, Katherine D. McMahon5,6†, Brett J. Baker1,7‡
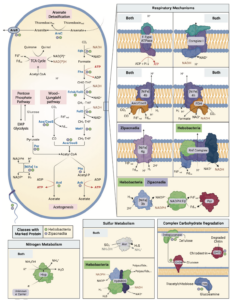
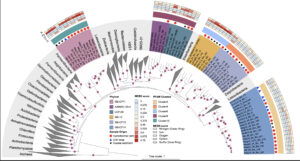
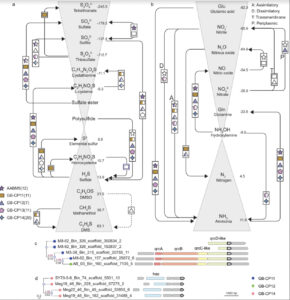
Ecology and evolution are distinct theories, but the short lifespans and large population sizes of microbes allow evolution to unfold along contemporary ecological time scales. To document this in a natural system, we collected a two-decade, 471-metagenome time series from a single site in a freshwater lake, which we refer to as the TYMEFLIES dataset. This massive sampling and sequencing effort resulted in the reconstruction of 30,389 metagenomic-assembled genomes (MAGs) over 50% complete, which dereplicated into 2,855 genotypes(>96% nucleotide sequence identity). We found both ecological and evolutionary processes occurred at seasonal time scales. There were recurring annual patterns at the species level in abundances, nucleotide diversities (π), and single nucleotide variant (SNV) profiles. During annual blooms, we observed both higher and lower nucleotide diversity, indicating that both ecological differentiation and competition drove evolutionary dynamics. Overlayed upon seasonal patterns, we observed long-term change in 20% of the species’ SNV profiles including gradual changes, step changes, and disturbances followed by resilience. Most abrupt changes occurred in a single species, suggesting evolutionary drivers are highly specific. Nevertheless, seven members of the abundant Nanopelagicaceae family experienced abrupt change in 2012, an unusually hot and dry year. This shift coincided with increased numbers of genes under selection involved in amino acid and nucleic acid metabolism, suggesting fundamental organic nitrogen compounds drive strain differentiation in the most globally abundant freshwater family. Overall, we observed seasonal and decadal trends in both interspecific ecological and intraspecific evolutionary processes. The convergence of microbial ecology and evolution on the same time scales demonstrates that understanding microbiomes requires a new unified approach that views ecology and evolution as a single continuum.