
A “behind the paper” article Brett wrote for the Nature Community website.



Baker Marine Microbial Ecology Lab
We use culture independent techniques (genomics, transcriptomics, and proteomics) to understand the ecology and evolution of microbial communities.
Our new paper in collaboration with Thijs Ettema’s lab describes a new model for the partnership that led to the origin of eukaryotes.
Summary
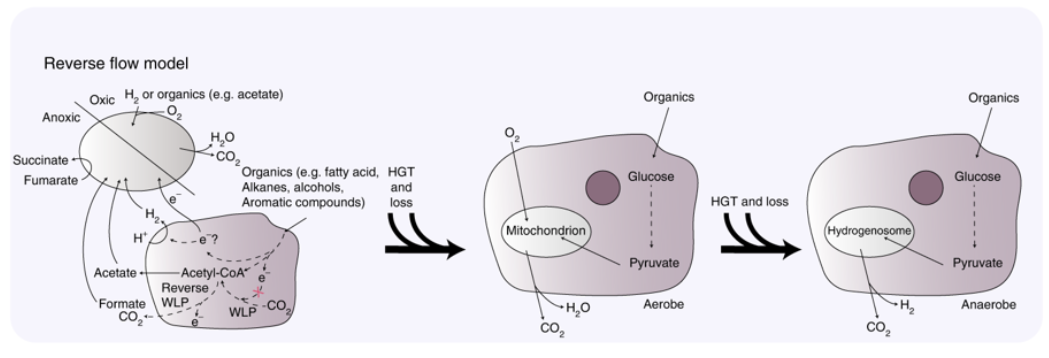
The origin of eukaryotes represents an unresolved puzzle in evolutionary biology. Current research suggests that eukaryotes evolved from a merger between a host of archaeal descent and an alphaproteobacterial endosymbiont. The discovery of the Asgard archaea, a proposed archaeal superphylum that includes Lokiarchaeota, Thorarchaeota, Odinarchaeota and Heimdallarchaeota suggested to comprise the closest archaeal relatives of eukaryotes, has helped to elucidate the identity of the putative archaeal host. Whereas Lokiarchaeota are assumed to employ a hydrogen-dependent metabolism, little is known about the metabolic potential of other members of the Asgard superphylum. We infer the central metabolic pathways of Asgard archaea using comparative genomics and phylogenetics to be able to refine current models for the origin of eukaryotes. Our analyses indicate that Thorarchaeota and Lokiarchaeota encode proteins necessary for carbon fixation via the Wood–Ljungdahl pathway and for obtaining reducing equivalents from organic substrates. By contrast, Heimdallarchaeum LC2 and LC3 genomes encode enzymes potentially enabling the oxidation of organic substrates using nitrate or oxygen as electron acceptors. The gene repertoire of Heimdallarchaeum AB125 and Odinarchaeumindicates that these organisms can ferment organic substrates and conserve energy by coupling ferredoxin reoxidation to respiratory proton reduction. Altogether, our genome analyses suggest that Asgard representatives are primarily organoheterotrophs with variable capacity for hydrogen consumption and production. On this basis, we propose the ‘reverse flow model’, an updated symbiogenetic model for the origin of eukaryotes that involves electron or hydrogen flow from an organoheterotrophic archaeal host to a bacterial symbiont.
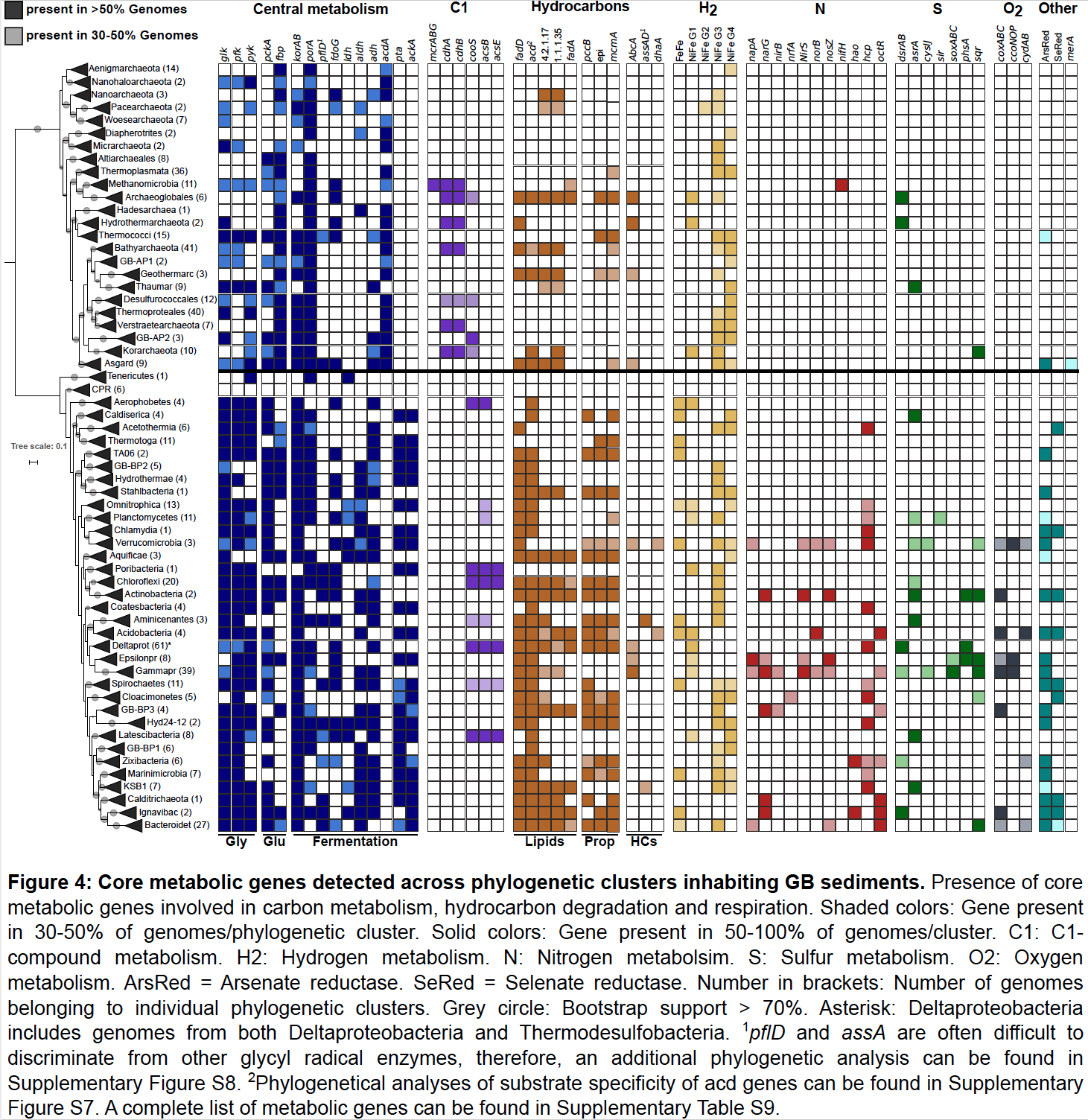
Microbes in Guaymas Basin (Gulf of California) hydrothermal sediments thrive on hydrocarbons and sulfur and experience steep, fluctuating temperature and chemical gradients. The functional capacities of communities inhabiting this dynamic habitat are largely unknown. Here, we reconstructed 551 genomes from hydrothermally influenced, and nearby cold sediments belonging to 56 phyla (40 uncultured). These genomes comprise 22 unique lineages, including five new candidate phyla. In contrast to findings from cold hydrocarbon seeps, hydrothermal-associated communities are more diverse and archaea dominate over bacteria. Genome-based metabolic inferences provide first insights into the ecological niches of these uncultured microbes, including methane cycling in new Crenarchaeota and alkane utilization in ANME-1. These communities are shaped by a high biodiversity, partitioning among nitrogen and sulfur pathways and redundancy in core carbon-processing pathways. The dynamic sediments select for distinctive microbial communities that stand out by expansive biodiversity, and open up new physiological perspectives into hydrothermal ecosystem function.


Diphthamide is a modified histidine residue which is uniquely present in archaeal and eukaryotic elongation factor 2 (EF-2), an essential GTPase responsible for catalyzing the coordinated translocation of tRNA and mRNA through the ribosome. In part due to the role of diphthamide in maintaining translational fidelity, it was previously assumed that diphthamide biosynthesis genes (dph) are conserved across all eukaryotes and archaea. Here, comparative analysis of new and existing genomes reveals that some archaea (i.e., members of the Asgard superphylum, Geoarchaea, and Korarchaeota) and eukaryotes (i.e., parabasalids) lack dph. In addition, while EF-2 was thought to exist as a single copy in archaea, many of these dph-lacking archaeal genomes encode a second EF-2 paralog missing key-residues required for diphthamide modification and for normal translocase function, perhaps suggesting functional divergence linked to loss of diphthamide biosynthesis. Interestingly, some Heimdallarchaeota previously suggested to be most closely related to the eukaryotic ancestor maintain dph genes and a single gene encoding canonical EF-2. Our findings reveal that the ability to produce diphthamide, once thought to be a universal feature in archaea and eukaryotes, has been lost multiple times during evolution, and suggest that anticipated compensatory mechanisms evolved independently.




Here is some of the press that covered the impacts of the storm on my lab, institute, lab members, and family.
