Asgard archaea illuminate the origin of eukaryotic cellular complexity

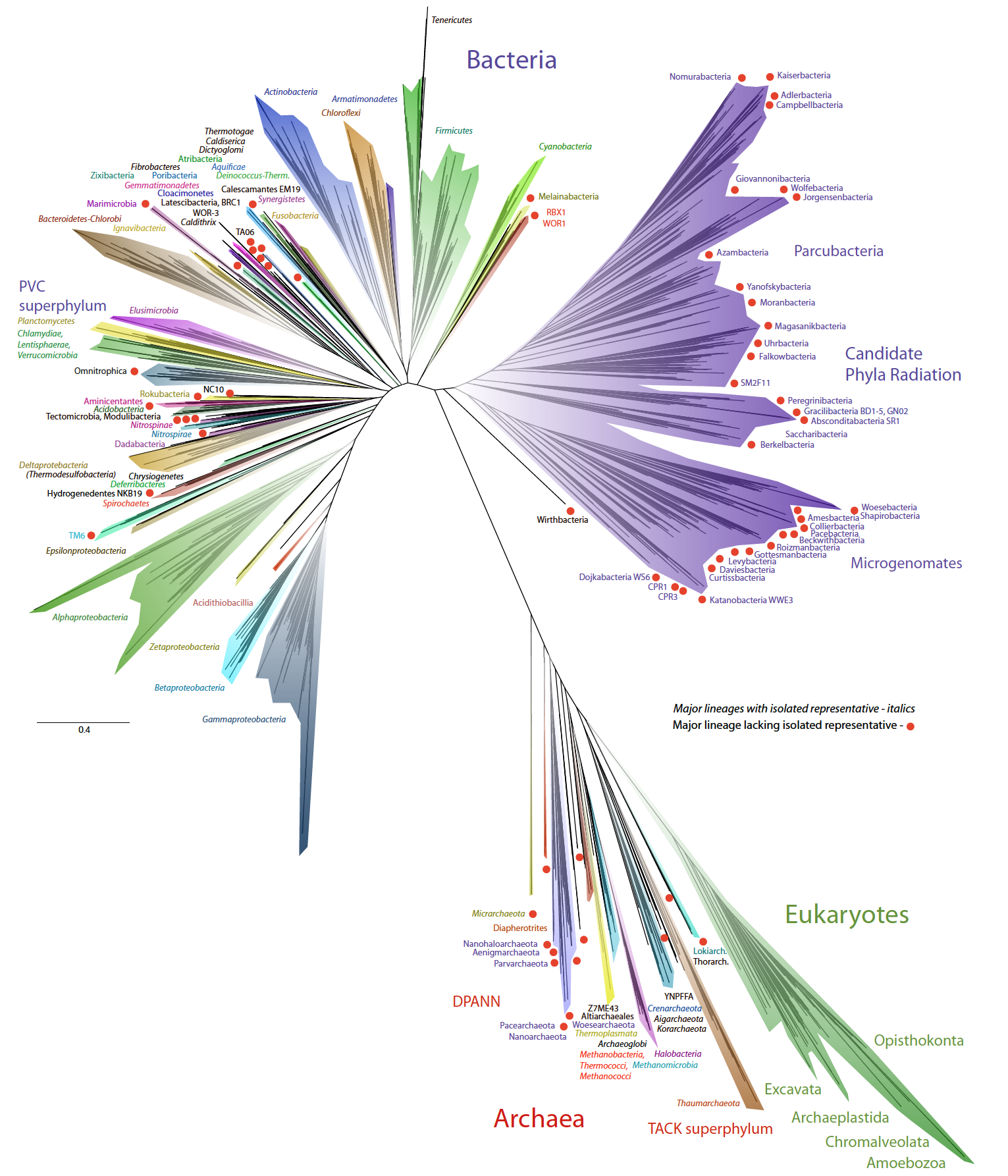
 This week our new paper describing the discovery of four archaea phyla that are related to eukaryotes was published in Nature. These phyla belong to the same branch of life and have been named after different Norse gods, Thor, Odin, Heimdall, and Loki. This is a collaboration with Thijs Ettema’s lab in Sweden. Last year we published the discovery of Thorarchaeota in ISME.
This week our new paper describing the discovery of four archaea phyla that are related to eukaryotes was published in Nature. These phyla belong to the same branch of life and have been named after different Norse gods, Thor, Odin, Heimdall, and Loki. This is a collaboration with Thijs Ettema’s lab in Sweden. Last year we published the discovery of Thorarchaeota in ISME.
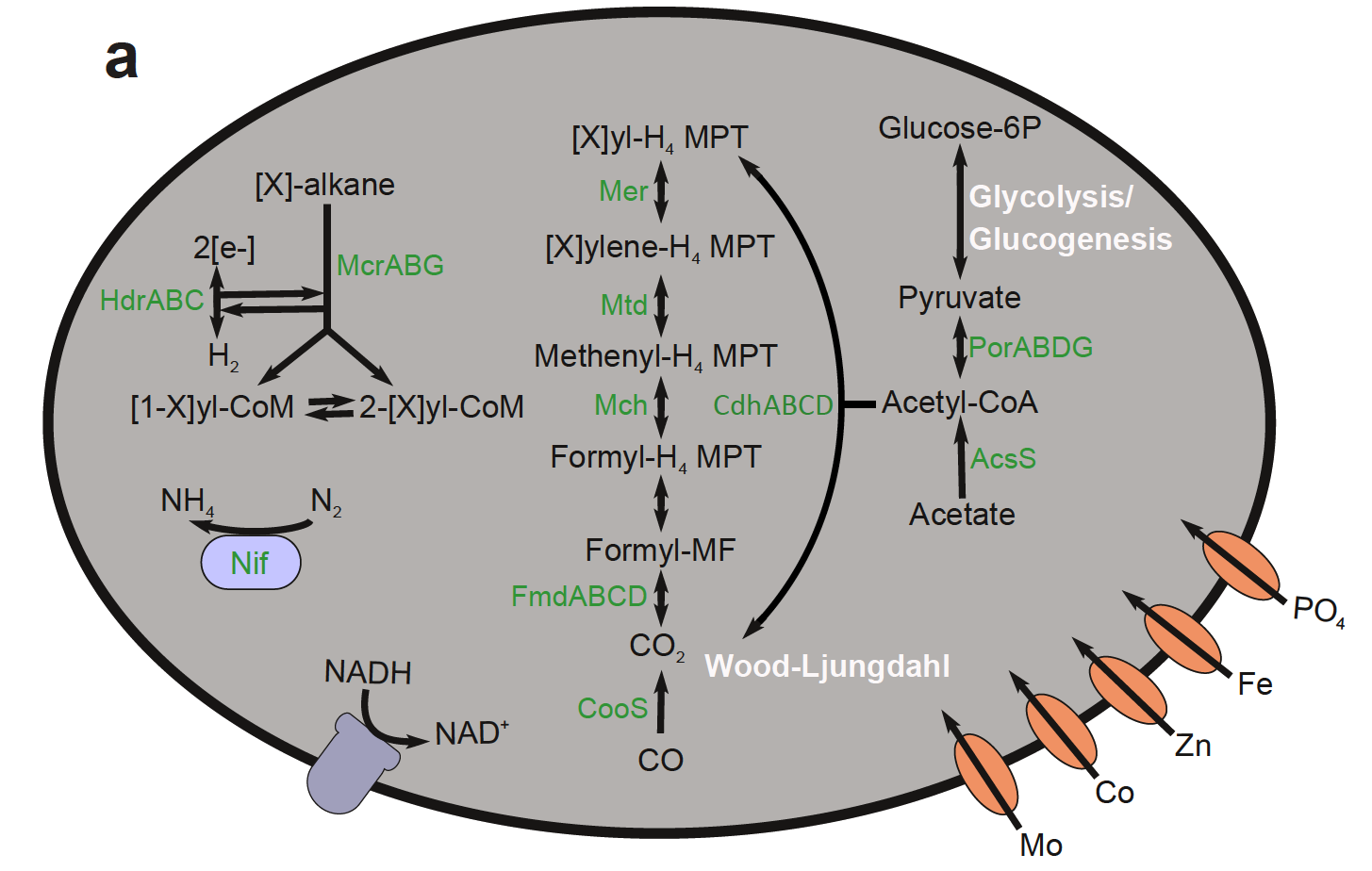
Genomic reconstruction of a novel, deeply branched sediment archaeal phylum with pathways for acetogenesis and sulfur reduction
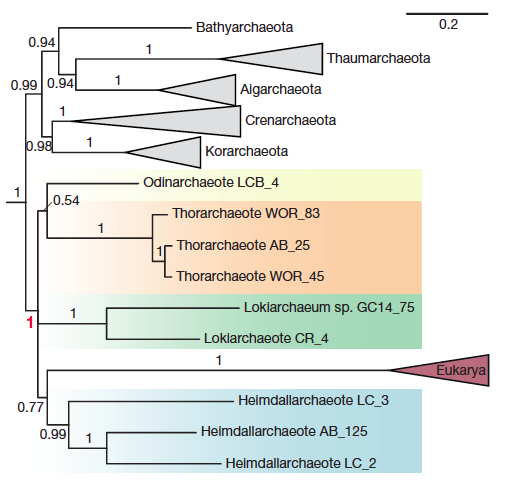
This paper adds two additional phyla, Odinarchaeota and Heimdallarchaeota. The focus of this paper is to further resolve the phylogenetic position of eukaryotes in this new superphylum. It also examines the presence of several new ESPs or eukaryotic signature proteins. These proteins were mostly thought to exist in eukaryotes, but these genomes contain a variety of them!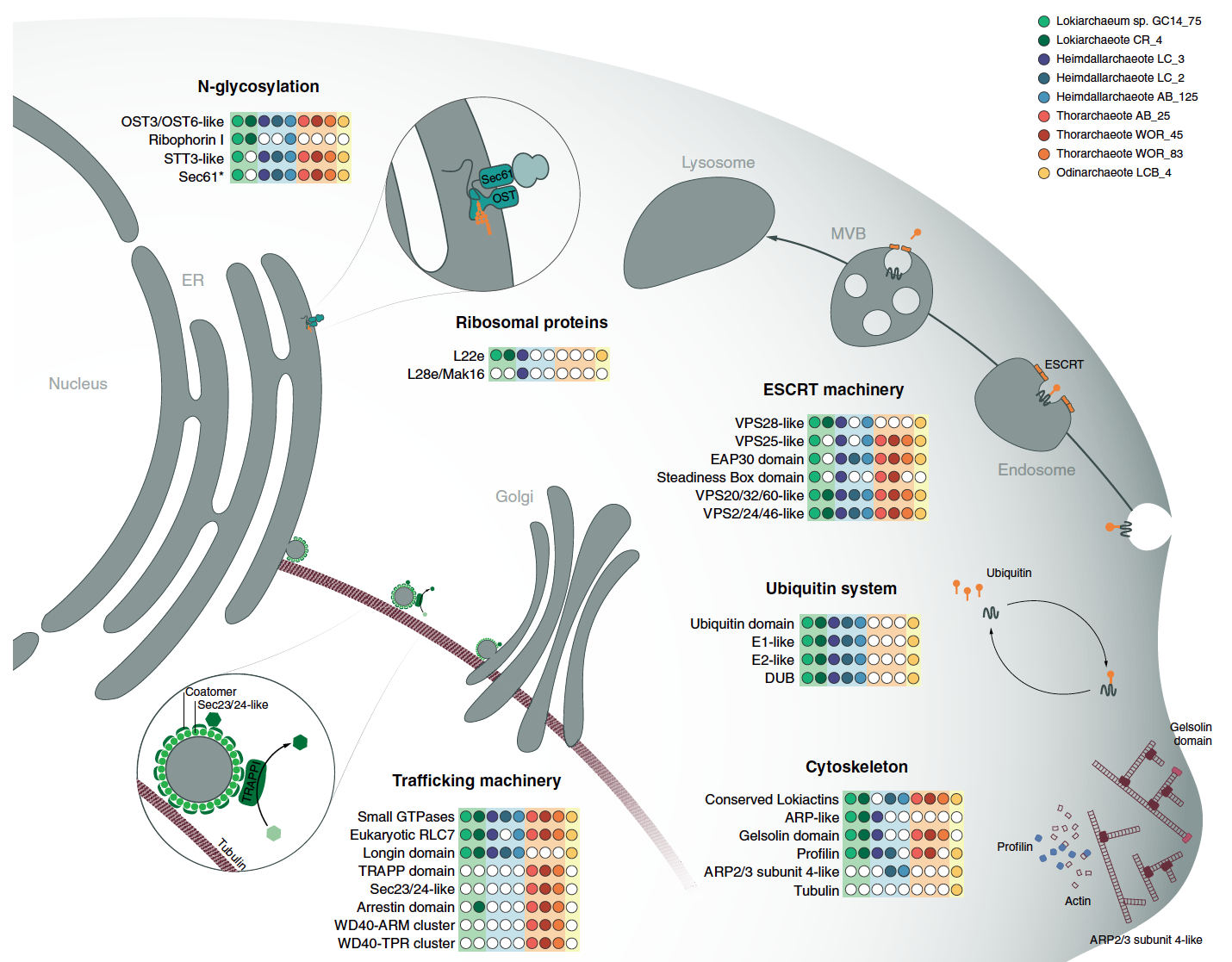
Press releases to accompany this study:
UT press release
The Atlantic article by Ed Yong
Uppsala press release


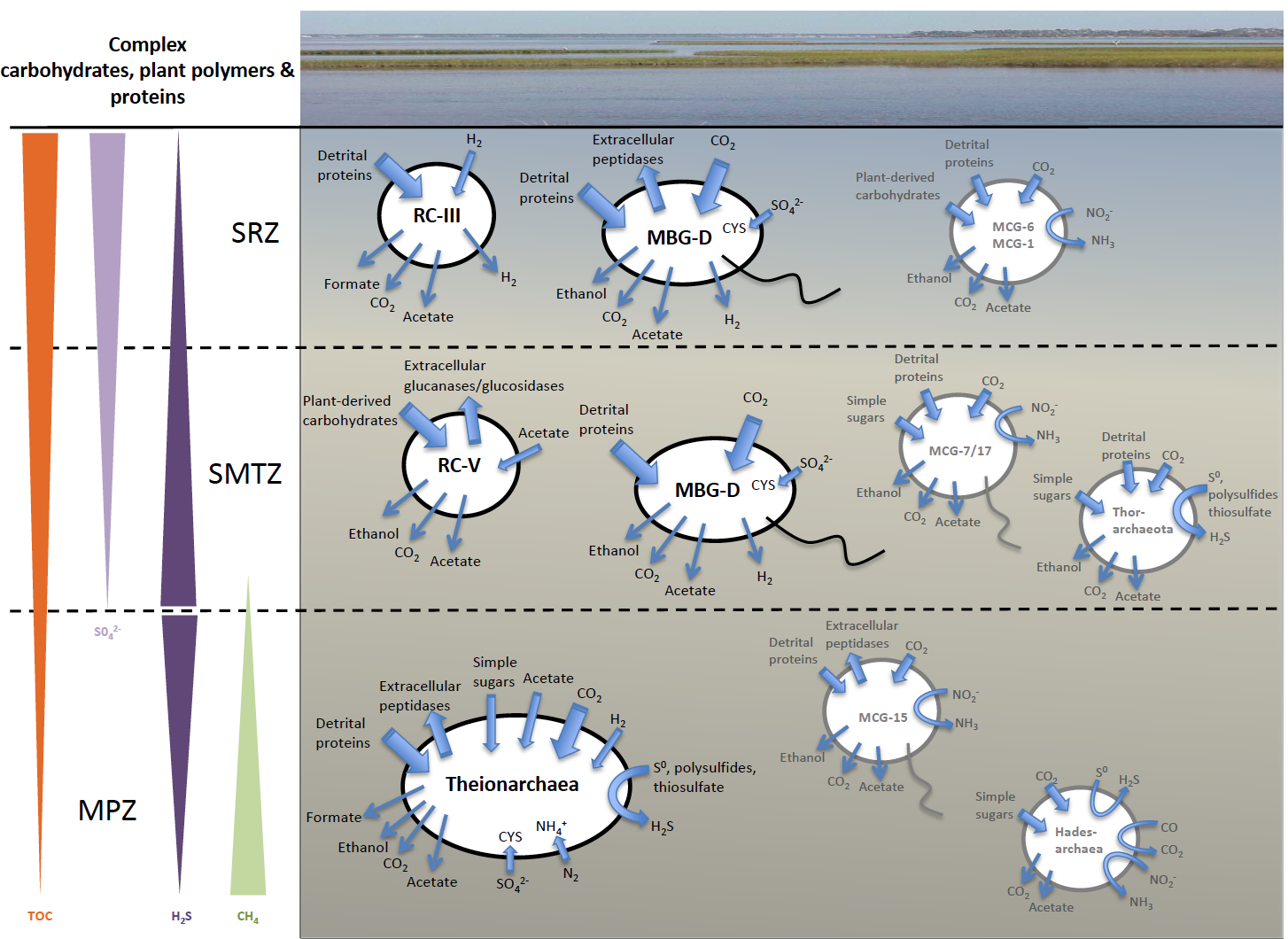

 This week our new paper describing the discovery of four archaea phyla that are related to eukaryotes was published in Nature. These phyla belong to the same branch of life and have been named after different Norse gods, Thor, Odin, Heimdall, and Loki. This is a collaboration with Thijs Ettema’s lab in Sweden. Last year we published the discovery of Thorarchaeota in ISME.
This week our new paper describing the discovery of four archaea phyla that are related to eukaryotes was published in Nature. These phyla belong to the same branch of life and have been named after different Norse gods, Thor, Odin, Heimdall, and Loki. This is a collaboration with Thijs Ettema’s lab in Sweden. Last year we published the discovery of Thorarchaeota in ISME.
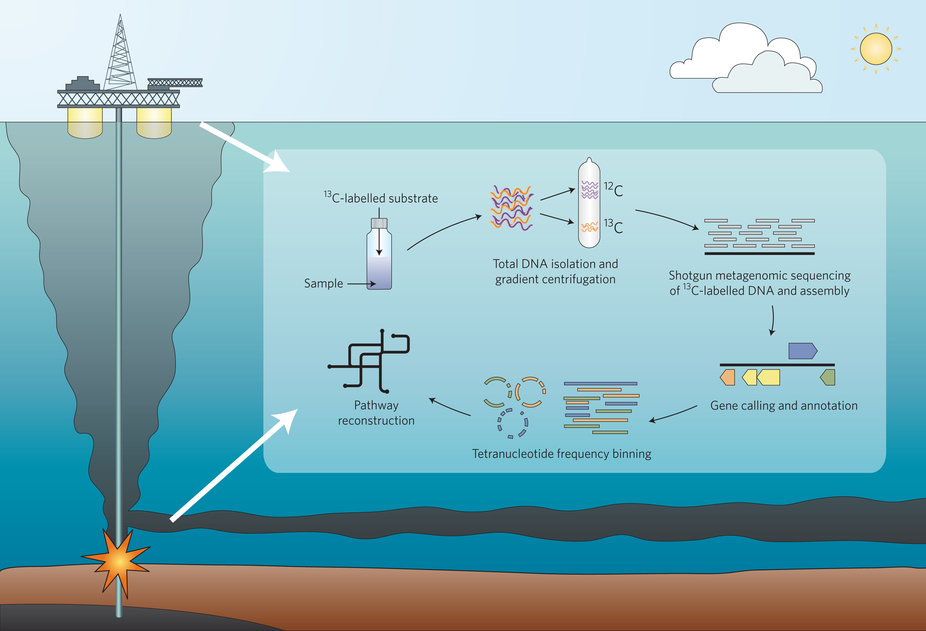 DNA-SIP metagenomic experimental strategy for the identification and characterization of hydrocarbon-degrading microorgansims from DWH oil spill deep-plume and surface-slick samples.
DNA-SIP metagenomic experimental strategy for the identification and characterization of hydrocarbon-degrading microorgansims from DWH oil spill deep-plume and surface-slick samples.