Here is some of the press that covered the impacts of the storm on my lab, institute, lab members, and family.
Nature – How labs are coping with Hurricane Harvey’s devastating floods
Science – A lab flees from Harvey: We were ‘just so damn lucky’


We use culture independent techniques (genomics, transcriptomics, and proteomics) to understand the ecology and evolution of microbial communities.
Here is some of the press that covered the impacts of the storm on my lab, institute, lab members, and family.

Congrats Nina and Kiley!
This paper details the genetic diversity of these sediments and describes genomes belonging to a uncultured archaeal group (GoM-Arc1) that contain novel pathways for hydrocarbon cycling, related to ANME (anaerobic methane oxidizers).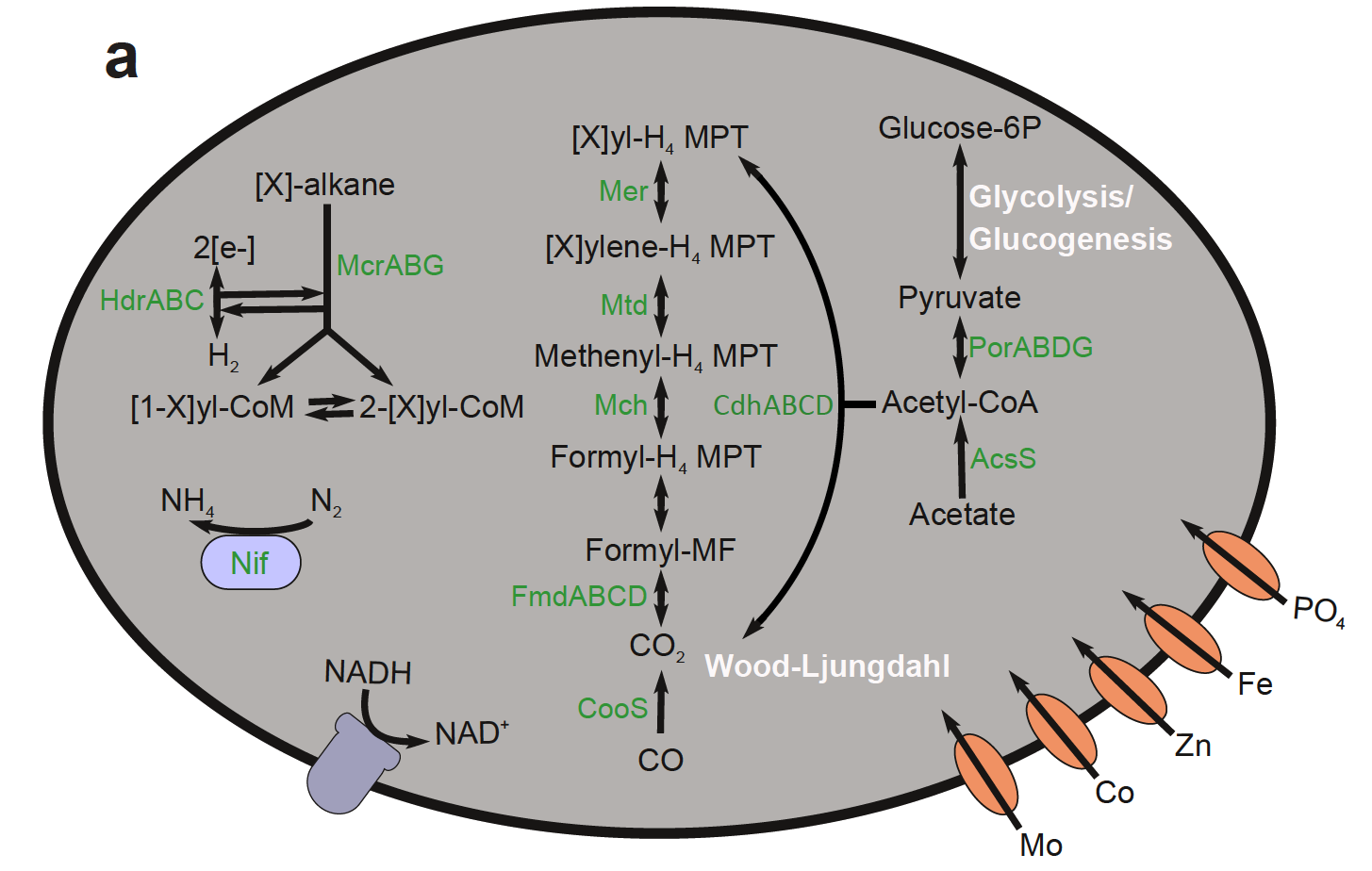

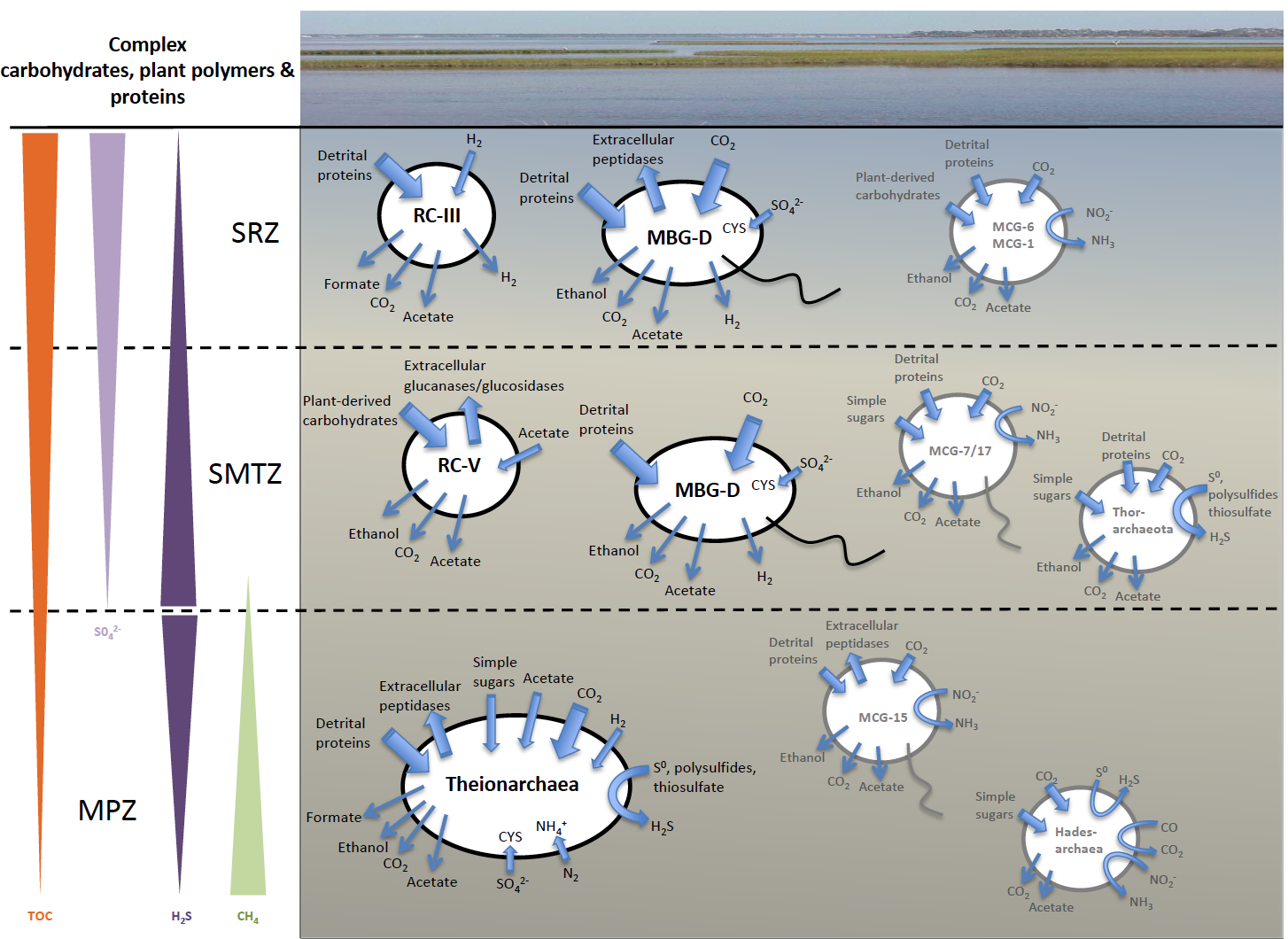
In addition to Theionarchaea, this new paper that appears in ISME Journal also details a variety of archaeal genomes there were obtained from the White Oak River Estuary in North Carolina. This digram summarizes the ecological roles we have inferred from these genomes. This is important because NONE of these lineages have been grown in a laboratory, so having their genomes has significantly advanced our understanding of what they do in nature.
Asgard archaea illuminate the origin of eukaryotic cellular complexity

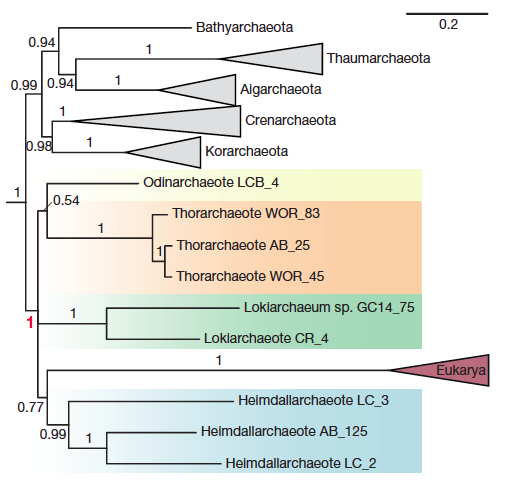 This week our new paper describing the discovery of four archaea phyla that are related to eukaryotes was published in Nature. These phyla belong to the same branch of life and have been named after different Norse gods, Thor, Odin, Heimdall, and Loki. This is a collaboration with Thijs Ettema’s lab in Sweden. Last year we published the discovery of Thorarchaeota in ISME.
This week our new paper describing the discovery of four archaea phyla that are related to eukaryotes was published in Nature. These phyla belong to the same branch of life and have been named after different Norse gods, Thor, Odin, Heimdall, and Loki. This is a collaboration with Thijs Ettema’s lab in Sweden. Last year we published the discovery of Thorarchaeota in ISME.
This paper adds two additional phyla, Odinarchaeota and Heimdallarchaeota. The focus of this paper is to further resolve the phylogenetic position of eukaryotes in this new superphylum. It also examines the presence of several new ESPs or eukaryotic signature proteins. These proteins were mostly thought to exist in eukaryotes, but these genomes contain a variety of them!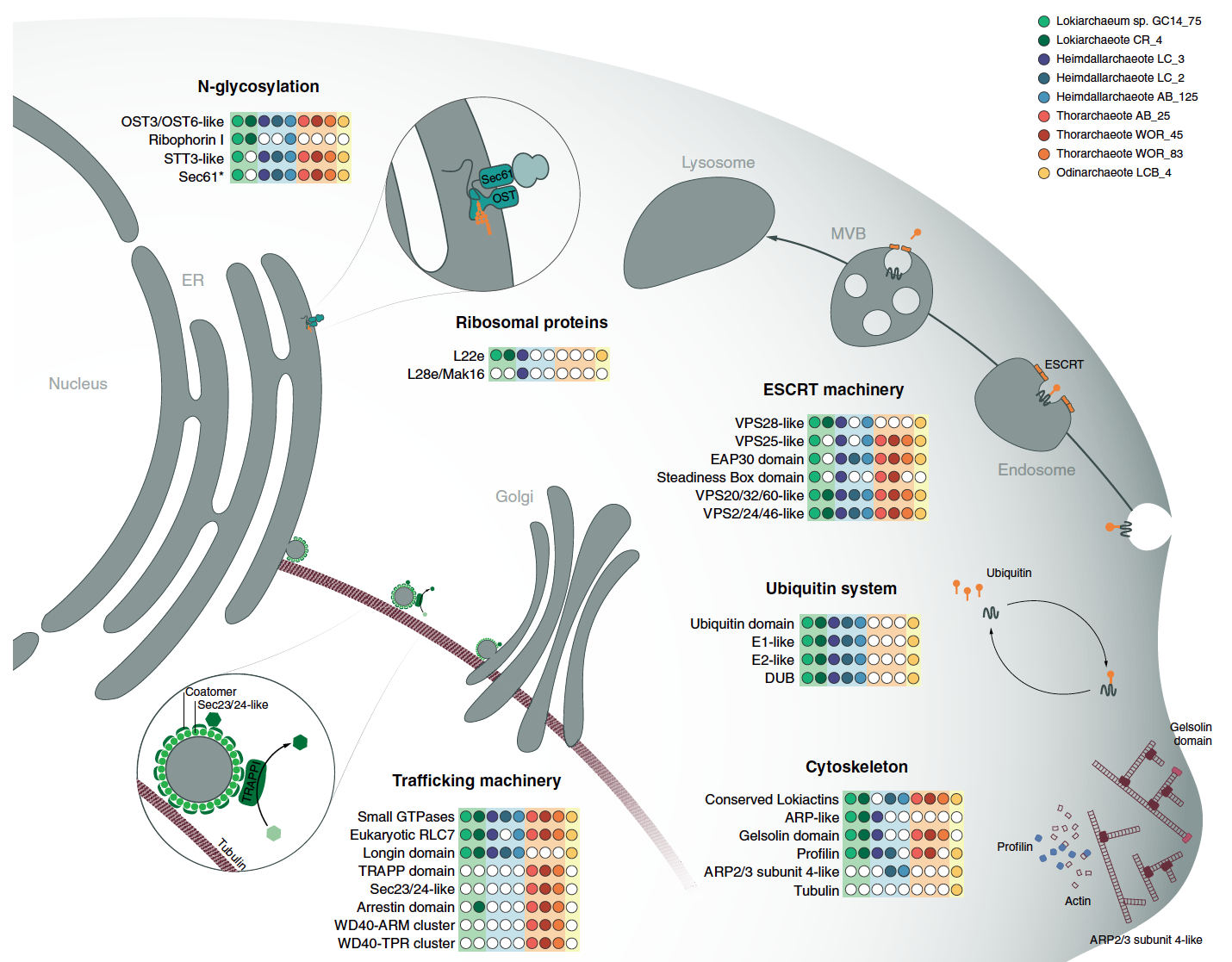
Press releases to accompany this study:
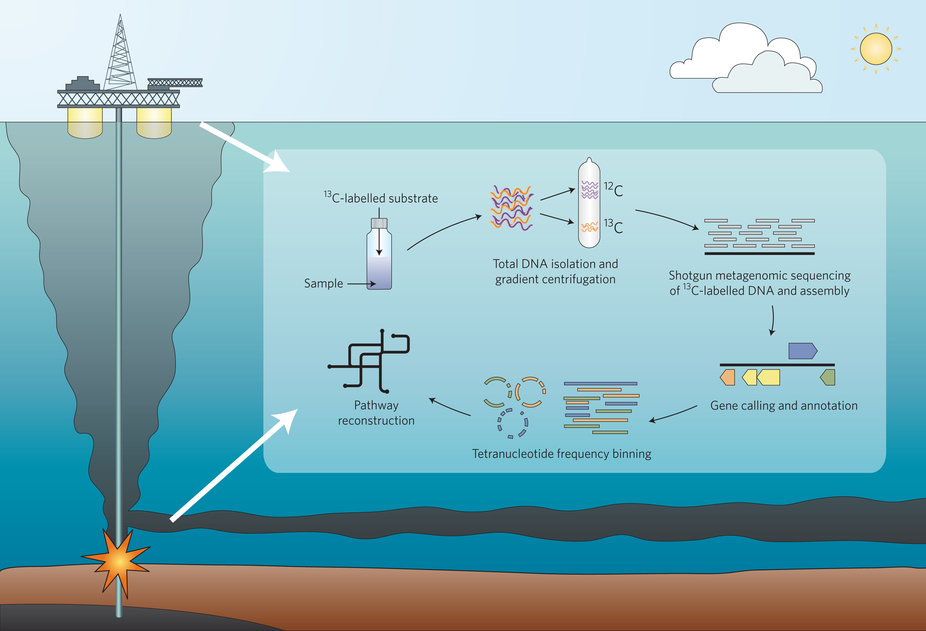 DNA-SIP metagenomic experimental strategy for the identification and characterization of hydrocarbon-degrading microorgansims from DWH oil spill deep-plume and surface-slick samples.
DNA-SIP metagenomic experimental strategy for the identification and characterization of hydrocarbon-degrading microorgansims from DWH oil spill deep-plume and surface-slick samples.
Through a collaboration with the Teske Lab (UNC), Tony Gutierrez (my 2015 REU student), and postdoc (Nina Dombrowski), several genomes were reconstructed from DNA-SIP experiments run on the DWH spill oil in 2010, (resulting in finding hydrocarbon degradation pathways in uncultured bacteria from the spill).
This cover of The National from Scotland.
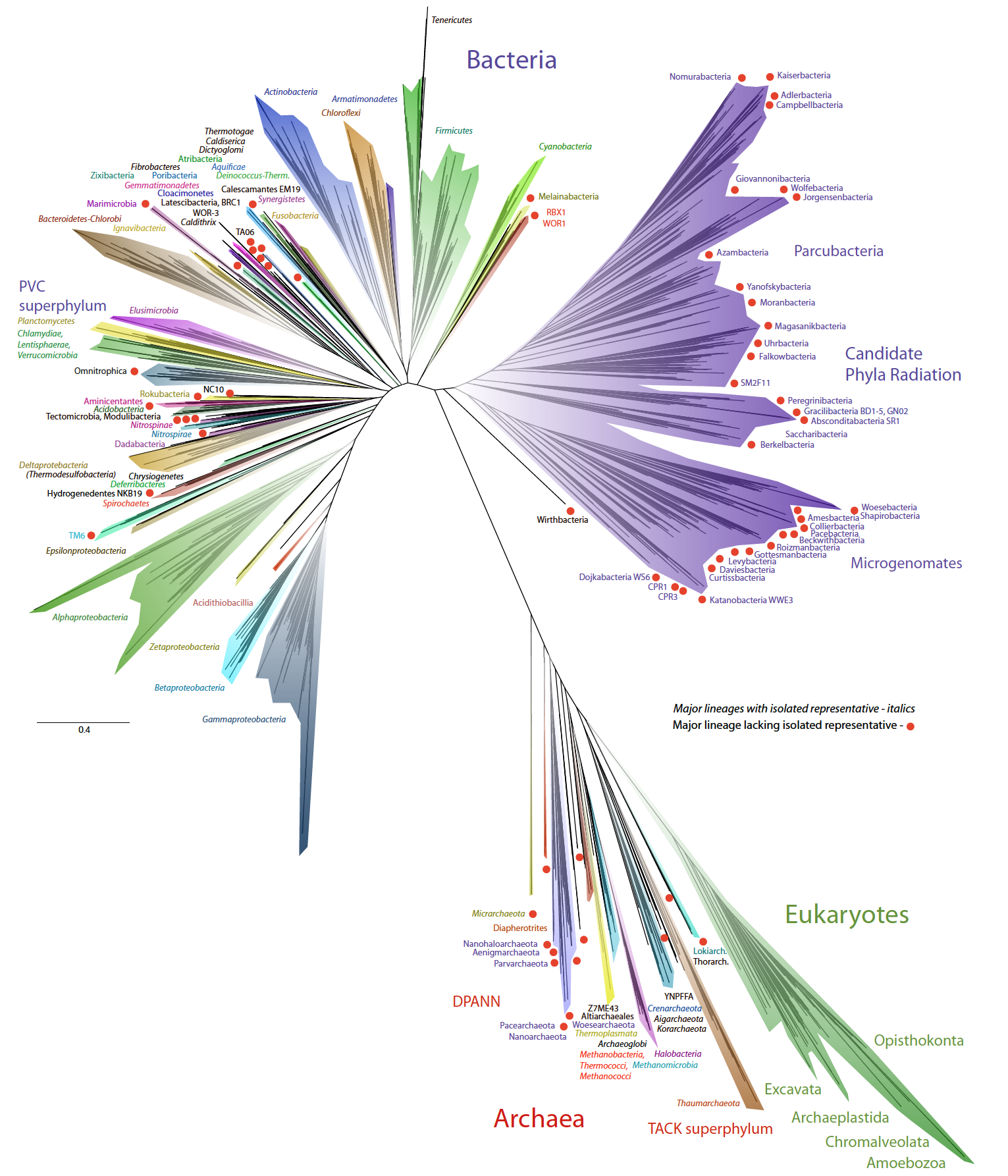
A new view of the tree of life
The rapid sequencing of new microbial genomes in recent years has left us wondering what all this new data has done to the tree of life. To address this, a new paper that presents the new view of life diversity in the genomic era has just been published in Nature Microbiology.
This paper is a collaboration with many people, namely Laura Hug and Jill Banfield (at UC Berkeley).