Huge congrats to Jess Sterling, who just successfully defended her PhD!
Jess has been a great lab member and researcher, who has made superb progress over the last 5 years. She joined the lab to work on immunity in marine systems, but ended up getting excited about how mitochondria might contribute to aging. She’s done side projects on bacterial communities in anchialine ecosystems, population genetics, and mitochondrial evolution of different animals, but her dissertation is titled “Discerning Mechanisms of Mitochondrial DNA (mtDNA) Mutation through Sequence Analysis”.
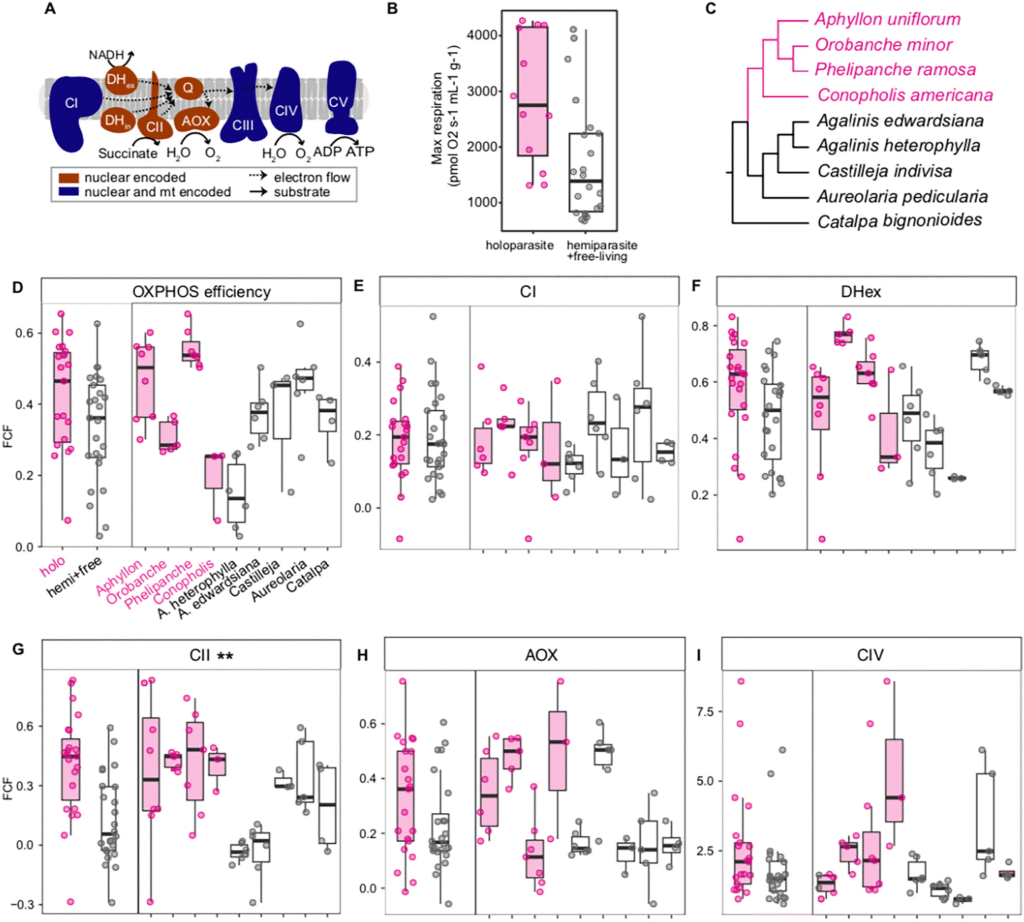
In her first chapter, she examined patterns of mtDNA sequence evolution in 4 groups of animals with quite variable lifespans to see if any mtDNA evolutionary patterns correlated with lifespan. She supported some previous findings showing that vertebrates with longer lifespans might have decreased mtDNA mutation rates, but ultimately she concluded that this was probably a methodological artifact and the pattern disappears when taking generation time into account. This story (and more) is in review now and should be published soon.

Her other projects made use of the model nematode C. elegans and several naturally occuring strains with variable lifespans. She characterized mitochondrial physiology among the strains and performed duplex sequencing to ask how mitochondrial mutations accumulated throughout the life of different strains. She’s wrapping up this work now and finding some really interesting conclusions, suggesting what’s been described in mammals might not apply to animals at large.
Finally, Jess has secured her “dream job” which she’ll be starting next Spring, working for the Boston Consulting Group (and staying based in Austin!).
Congrats again Jess!!!