Oxygen metabolism in descendants of the archaeal-eukaryotic ancestor
https://doi.org/10.1038/s41586-026-10128-z
Article by Carl Zimmer in the New York Times
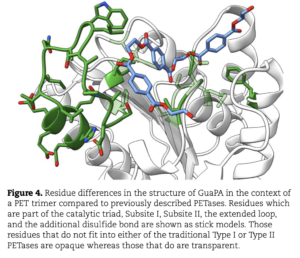
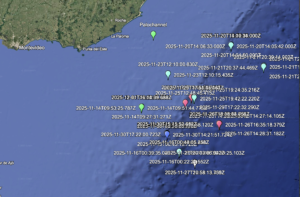
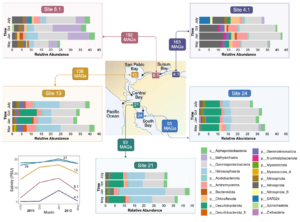
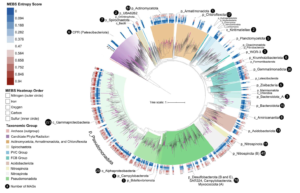
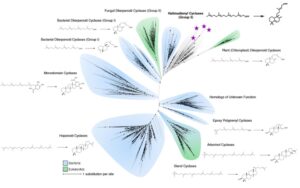
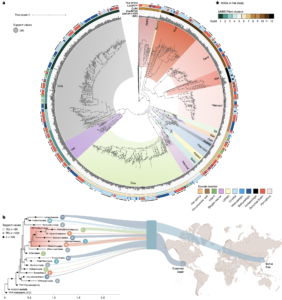
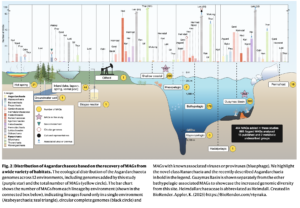
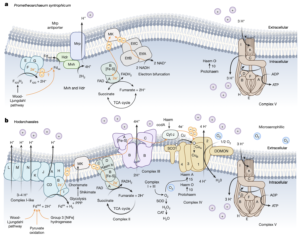
Asgard archaea were pivotal in the origin of complex cellular life1. Heimdallarchaeia (a class within the phylum Asgardarchaeota) are inferred to be the closest relatives of eukaryotes. Limited sampling of these archaea constrains our understanding of their ecology and evolution, including their role in eukaryogenesis. Here we use massive DNA sequencing of marine sediments to obtain 404 Asgardarchaeota metagenome-assembled genomes, including 136 new Heimdallarchaeia and several novel lineages. Analyses of their global distribution revealed they are widespread in marine environments, and many are enriched in variably oxygenated coastal sediments. Detailed metabolic reconstructions and structural predictions suggest that Heimdallarchaeia form metabolic guilds that are distinct from other Asgardarchaeota. These archaea encode hallmark proteins of an aerobic lifestyle, including electron transport chain complex (IV), haem biosynthesis and reactive oxygen species detoxification. Heimdallarchaeia also encode novel clades of respiratory membrane-bound hydrogenases with additional Complex I-like subunits, which potentially increase proton-motive force generation and ATP synthesis. Thus, we propose an updated Heimdallarchaeia-centric model of eukaryogenesis in which hydrogen production and aerobic respiration may have been present in the Asgard-eukaryotic ancestor. This expanded catalogue of Asgard archaeal genomic diversity suggests that bioenergetic factors influenced eukaryogenesis and constitutes a valuable resource for investigations into the origins and evolution of cellular complexity.
UT press release