It’s been great to have Sophie working in the lab!


We use culture independent techniques (genomics, transcriptomics, and proteomics) to understand the ecology and evolution of microbial communities.
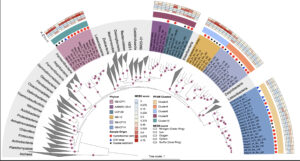
Microbes in marine sediments play crucial roles in global carbon and nutrient cycling. However, our understanding of microbial diversity and physiology on the ocean floor is limited. Here, we use phylogenomic analyses of thousands of metagenome-assembled genomes (MAGs) from coastal and deep-sea sediments to identify 55 MAGs that are phylogenetically distinct from previously described bacterial phyla. We propose that these MAGs belong to 4 novel bacterial phyla (Blakebacterota, Orphanbacterota, Arandabacterota, and Joyebacterota) and a previously proposed phylum (AABM5-125-24), all of them within the FCB superphylum. Comparison of their rRNA genes with public databases reveals that these phyla are globally distributed in different habitats, including marine, freshwater, and terrestrial environments. Genomic analyses suggest these organisms are capable of mediating key steps in sedimentary biogeochemistry, including anaerobic degradation of polysaccharides and proteins, and respiration of sulfur and nitrogen. Interestingly, these genomes code for an unusually high proportion (~9% on average, up to 20% per genome) of protein families lacking representatives in public databases. Genes encoding hundreds of these protein families co-localize with genes predicted to be involved in sulfur reduction, nitrogen cycling, energy conservation, and degradation of organic compounds. Our findings advance our understanding of bacterial diversity, the ecological roles of these bacteria, and potential links between novel gene families and metabolic processes in the oceans.
Genomes of six viruses that infect Asgard archaea from deep-sea sediments
Read more in the UT press release here.

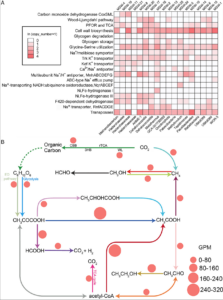
Reduced substrates produced by the serpentinization reaction under hydration of olivine may have fuelled biological processes on early Earth. To understand the adaptive strategies and carbon metabolism of the microbes in the serpentinizing ecosystems, we reconstructed 18 draft genomes representing dominant species of Omnitrophicaeota, Gammaproteobacteria and Methanobacteria from the Manleluag serpentinizing spring in Zambales, Philippines (hyperalkaline and rich in methane and hydrogen). Phylogenomics revealed that two genomes were affiliated with a candidate phylum NPL-UPA2 and the references of all our genomes were derived from ground waters, hot springs and the deep biosphere. C1 metabolism appears to be widespread as most of the genomes code for methanogenesis, CO oxidation and CO2 fixation. However, likely due to the low CO2concentration and election acceptors, the biomass in the spring was extremely low (<103 cell/ml). Various Na+ and K+ transporters and Na+-driving ATPases appear to be encoded by these genomes, suggesting that nutrient acquisition, bioenergetics and normal cytoplasmic pH were dependent on Na+ and K+ pumps. Our results advance our understanding of the metabolic potentials and bioenergetics of serpentinizing springs and provide a framework of the ecology of early Earth.
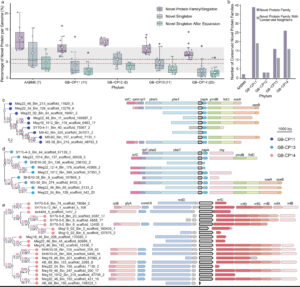
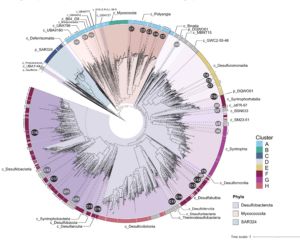
Deltaproteobacteria, now proposed to be the phyla Desulfobacterota, Myxococcota, and SAR324, are ubiquitous in marine environments and play essential roles in global carbon, sulfur, and nutrient cycling. Despite their importance, our understanding of these bacteria is biased towards cultured organisms. Here we address this gap by compiling a genomic catalog of 1,792 genomes, including 402 newly reconstructed and characterized metagenome-assembled genomes (MAGs) from coastal and deep-sea sediments. Phylogenomic analyses reveal that many of these novel MAGs are uncultured representatives of Myxococcota and Desulfobacterota that are understudied. To better characterize Deltaproteobacteria diversity, metabolism, and ecology, we clustered ~1,500 genomes based on the presence/absence patterns of their protein families. Protein content analysis coupled with large-scale metabolic reconstructions separates eight genomic clusters of Deltaproteobacteria with unique metabolic profiles. While these eight clusters largely correspond to phylogeny, there are exceptions where more distantly related organisms appear to have similar ecological roles and closely related organisms have distinct protein content. Our analyses have identified previously unrecognized roles in the cycling of methylamines and denitrification among uncultured Deltaproteobacteria. This new view of Deltaproteobacteria diversity expands our understanding of these dominant bacteria and highlights metabolic abilities across diverse taxa.
The start of it all (from The Texas Scientist magazine)
Dinosaurs, daisies, starfish and humans are all made up of cells containing tiny biological hitchhikers. In all complex life, each cell has an energy-producing organelle that has its own DNA and looks and acts suspiciously like bacteria. In animals, they’re called mitochondria; in plants, they’re called chloroplasts.
How they got there is an open question and one researchers like Brett Baker, a faculty member in The University of Texas at Austin Department of Marine Science, explore. Baker thinks the answer may go something like this:
About 2 billion years ago, a microbe called an archaeon gobbled up a free-floating bacterium, and it worked well for both parties. The bacterium provided new energy sources to the archaeon, which in turn provided safety and nutrients.

Early Archaeon
Over time the two previously free-living microbes spawned a single, hybrid offspring — the first eukaryote.
All complex, multicellular life on the planet evolved from this new lifeform.
Baker and his colleagues have uncovered evidence that a recently discovered group of microbes, called the Asgard archaea, were the original hosts to give rise to all eukaryotes, including us humans. With support from the Moore-Simons Project, the scientists are now investigating which Asgards are most closely related to eukaryotes and further exploring their physiological interactions and cellular structure.