PEER-REVIEWED PUBLICATIONS
2024
81. A. Cordova, B. Niese, P. Sweet, P. Kamat, J. M. Phillip, V. Gordon, and L. M. Contreras. “Geometric changes in the nucleoids of Deinococcus radiodurans reveal involvement of new proteins in recovery from ionizing radiation.” (Submitted). Preprint available from: https://www.biorxiv.org/content/10.1101/2024.01.17.576117v1.
80. P. Sweet, M. Burroughs, S. Jang, and L. M. Contreras. “TolRad: A model for predicting radiation tolerance using Pfam annotations identifies novel radiosensitive bacterial species from reference genomes and MAGs.” (Submitted). Preprint available from: https://biorxiv.org/cgi/content/short/2023.11.02.562514v1.
79. L. G. Miller, W. Kim, S. Schowe, K. Taylor, R. Han, V. Jain, R. Park, M. Sherman, J. Fang, H. Ramirez, A. Ellington, P. Tamamis, M. J. E. Resendiz, Y. J. Zhang, and L. M. Contreras. “Selective 8-oxo-rG stalling occurs in the catalytic core of polynucleotide phosphorylase (PNPase) during degradation.” (In Press).
78. A. M. Rojano-Nisimura, T. R. Simmons, A. N. Leistra, M. K. Mihailovic, R. Buchser, A. M. Ekdahl, G. Partipilo, I. Joseph, N. C. Curtis, and L. M. Contreras. “CsrA Shows Selective Regulation of sRNA-mRNA Networks.” (Accepted). Preprint available from: https://www.biorxiv.org/content/10.1101/2023.03.29.534774v1.
77. A. M. Rojano-Nisimura, K. B. Grismore, J. S. Ruzek, J. L. Avila and L. M. Contreras. “The Post-Transcriptional Regulatory Protein CsrA Amplifies Its Targetome through Direct Interactions with Stress-Response Regulatory Hubs: The EvgA and AcnA Cases.” Microorganisms 12, 636 (2024). Available from: https://www.mdpi.com/2076-2607/12/4/636.
76. K. E. Taylor, L. G. Miller, and L.M. Contreras. “RNA-binding proteins that preferentially interact with 8-oxoG-modified RNAs: our current understanding.” Biochem. Soc. Trans. (2024). Available from: https://doi.org/10.1042/BST20230254.
75. S.M. Engels, P. Kamat, G.S. Pafilis, Y. Li, A. Agrawal, D.J. Haller, J.M. Phillip, and L. M. Contreras. “Particulate matter composition drives differential molecular and morphological responses in lung epithelial cells.” PNAS Nexus 3, 1-15 (2024). Available from: https://doi.org/10.1093/pnasnexus/pgad415.
2023
74. L. Miller, M. Demny, P. Tamamis, and L. M. Contreras. “Characterization of epitranscriptome reader proteins experimentally and in silico: current knowledge and future perspectives beyond the YTH domain.” Comput. Struct. Biotechnol. J. 21, 3541–3556 (2023). Available from: https://doi.org/10.1016/j.csbj.2023.06.018.
73. R. Buchser, P. Sweet, A. Anantharaman, and L. M. Contreras. “RNAs as Sensors of Oxidative Stress in Bacteria.” Annual Review of Chemical and Biomolecular Engineering. 14, 265-281 (2023). Available from: https://doi.org/10.1146/annurev-chembioeng-101121-070250.
72. P. Sweet, J. Blacutt, V. Gordon, and L. M. Contreras. “Exposure of Shewanella oneidensis MR-1 to sub-lethal doses of ionizing radiation triggers short-term SOS activation and longer-term prophage activation.” Applied and Environmental Biology. Appl Environ Microb e01716-22 (2023). Available from: https://journals.asm.org/doi/10.1128/aem.01716-22.
2022
71. R. Han, J. Jiang, J. Fang, and L. M. Contreras. “PNPase and RhlB Interact and Reduce the Cellular Availability of Oxidized RNA in Deinococcus radiodurans.” Microbiology Spectrum 10, e02140-22 (2022). Available from: https://doi.org/10.1128/spectrum.02140-22.
70. A. M. Ekdahl, A. M. Rojano-Nisimura, and L. M. Contreras. “Engineering Toehold-Mediated Switches for Native RNA Detection and Regulation in Bacteria.” J Mol Bio 434, 18, 167689 (2022). Available from: https://doi.org/10.1016/j.jmb.2022.167689.
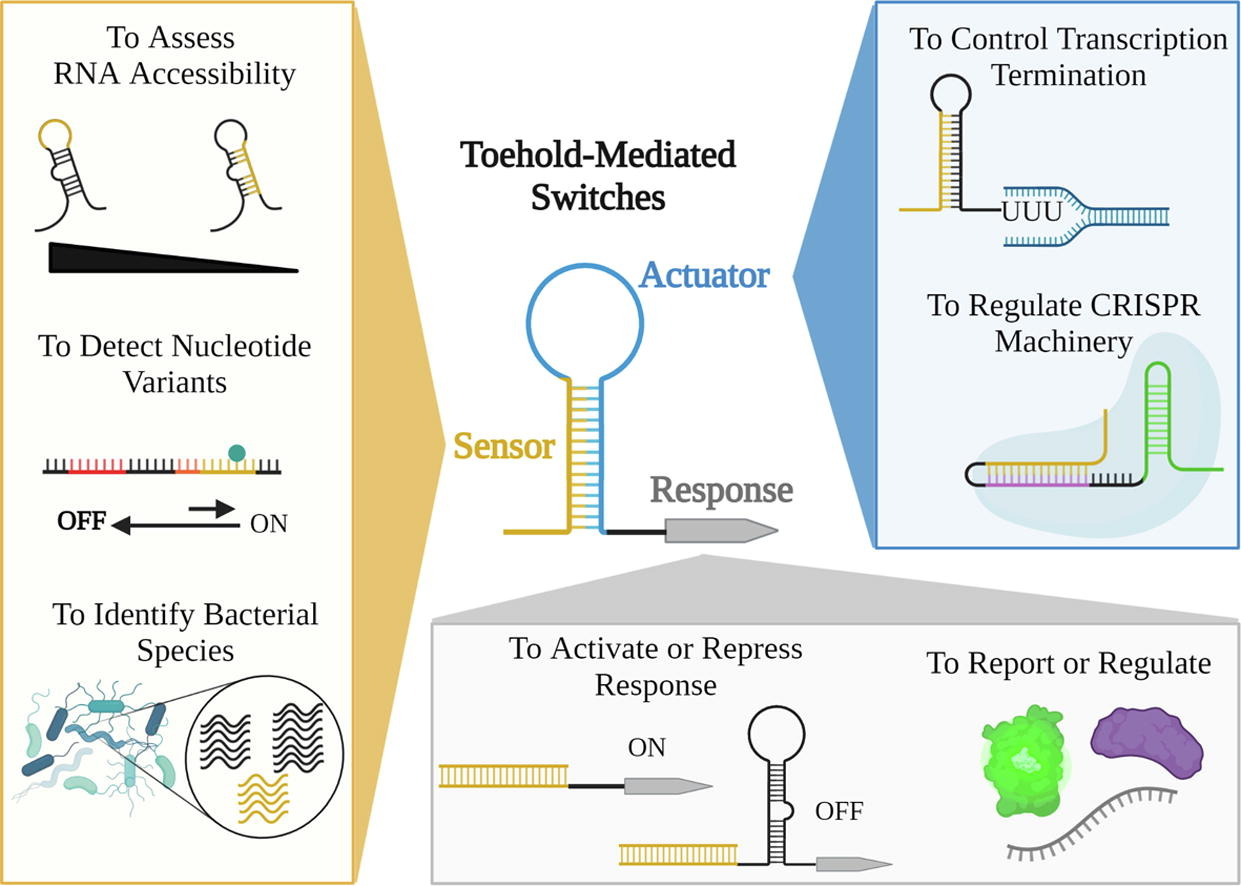
69. T. R. Simmons, A. D. Ellington, and L. M. Contreras. “RNP-Based Control Systems for Genetic Circuits in Synthetic Biology Beyond CRISPR.” Riboregulator Design and Analysis. Methods Mol Biology 1-31 (2022). Available from: https://doi.org/10.1007/978-1-0716-2421-0_1.
68. A. Hillsley, M. Santoso, S.M. Engels, K.N. Halwachs, L.M. Contreras, and A.M. Rosales. “A Strategy to Quantify Myofibroblast Activation on a Continuous Spectrum.” Sci Rep 12, 12239 (2022). Available from: https://doi.org/10.1038/s41598-022-16158-7.
2021
67. M. Chandler, B. Johnson, E. Khisamutdinov, M. A. Dobrovolskaia, J. Sztuba-Solinska, A. K. Salem, K. Breyne, R. Chammas, N. G. Walter, L. M. Contreras, P. Guo, and K. A. Afonin. “The International Society of RNA Nanotechnology and Nanomedicine (ISRNN): The Present and Future of the Burgeoning Field.” ACS Nano 15, 16957-16973 (2021). Available from: https://doi.org/10.1021/acsnano.0c10240.
66. M. R. Burroughs, J. C. Gonzalez-Rivera, A. Cordova, and L. M. Contreras. “Experimental and Computational Methods for Guiding Identification and Characterization of Epitranscriptome Proteins.” RNA Technologies 593–632 (2021). Available from: https://doi.org/10.1007/978-3-030-71612-7_22.
65. A. Chen, J. Hernández-Vargas, R. Han, O. Cortazar-Martínez, N. Gonzalez, S. Patel, B. Keitz, G. Luna-Barcenas, and L. M. Contreras. “Small RNAs as a New Platform for Tuning the Biosynthesis of Silver Nanoparticles for Enhanced Material and Functional Properties.” ACS Applied Materials & Interfaces (2021). Available from: https://doi.org/10.1021/acsami.1c07400.
64. M. Mihailovic, A. Ekdahl, A. Chen, A. Leistra, B. Li, J. González Martínez, M. Law, C. Ejindu, É. Massé, P. Freddolino and L. M. Contreras. “Uncovering Transcriptional Regulators and Targets of sRNAs Using an Integrative Data-Mining Approach: H-NS-Regulated RseX as a Case Study.” Frontiers in Cellular and Infection Microbiology 11, 696533 (2021). Available from: https://doi.org/10.3389/fcimb.2021.696533.
63. J.K. Villa, R. Han, C-H. Tsai, A. Chen, P. Sweet, G. Franco, R. Vaezian, R. Tkavc, M.J. Daly and L.M. Contreras. “A small RNA regulates pprM, a modulator of pleiotropic proteins promoting DNA repair, in Deinococcus radiodurans under ionizing radiation.” Scientific Reports 11, 12949 (2021). Available from: https://doi.org/10.1038/s41598-021-91335-8.
62. M. Sherman, S. Sandeep, and L. M. Contreras. “The Tryptophan-Induced tnaC Ribosome Stalling Sequence Exposes High Amino Acid Cross-Talk That Can Be Mitigated by Removal of NusB for Higher Orthogonality.” ACS Synthetic Biology (2021). Available from: https://doi.org/10.1021/acssynbio.0c00547.
2020
61. R. Han, J. Fang, J. Jiang, E. K. Gaidamakova, R. Tkavc, M. J. Daly and L.M. Contreras. “Signal Recognition Particle RNA Contributes to Oxidative Stress Response in Deinococcus radiodurans by Modulating Catalase Localization.” Frontiers in Microbiology 11, 613571 (2020). Available from: https://doi.org/10.3389/fmicb.2020.613571.
60. J.C. Gonzalez-Rivera, M.W. Sherman , D.S. Wang, J.C.L. Chuvalo-Abraham, L. Hildebrandt Ruiz, and L.M. Contreras. “RNA oxidation in chromatin modification and DNA-damage response following exposure to formaldehyde.” Scientific Reports 10, 16545 (2020). Available from: https://www.nature.com/articles/s41598-020-73376-7.
59. L.M. Contreras, J.C. Gonzalez-Rivera, K.C. Baldridge, D.S. Wang, J.C.L. Chuvalo-Abraham, and L. Hildebrandt Ruiz. “Understanding the Functional Impact of VOC-Ozone Mixtures on the Chemistry of RNA in Epithelial Lung Cells.” Res Rep Health Eff Inst, (201):201 (2020). Available from: https://pubmed.ncbi.nlm.nih.gov/32845096/.
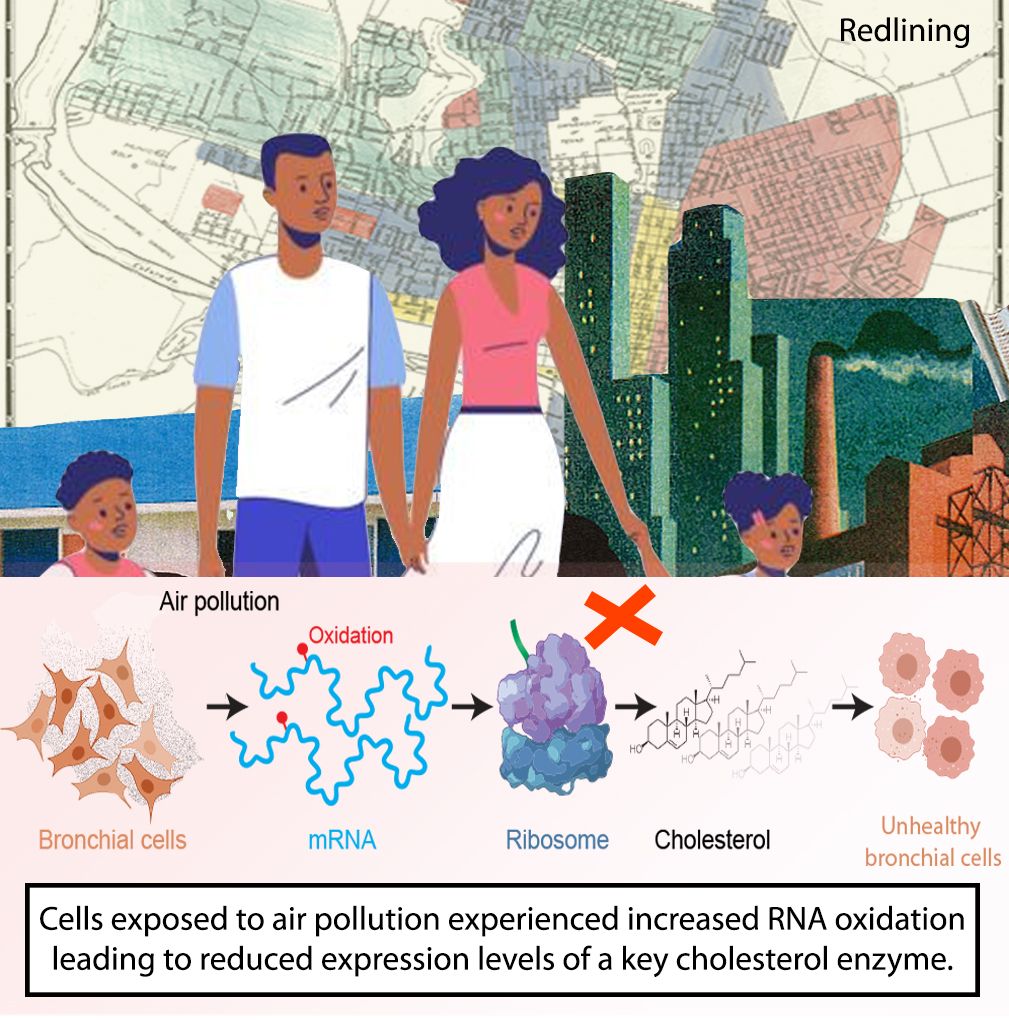
58. J.C. Gonzalez-Rivera, K.C. Baldridge, D.S. Wang, K.H. Patel, J.C.L. Chuvalo-Abraham, L. Hildebrandt Ruiz, and L.M. Contreras. “Post-transcriptional air pollution oxidation to the cholesterol biosynthesis pathway promotes pulmonary stress phenotypes” Communications Biology 3, 392 (2020). Available from: doi.org/10.1038/s42003-020-01118-6.
57. A.J. Gimenez, F. Medellin-Rodriguez, L.M. Contreras, J.M. Lopez-Romero, I. Sanchez, F. Villasenor-Ortega, G. Luna-Barcenas. “Turbidimetry by image degradation analysis” IEEE Transactions on Instrumentation and Measurement (2020). Available from: doi.org/10.1109/TIM.2020.2985902.
56. R. Han, K. Haning, J.C. Gonzalez-Rivera, Y. Yang, R. Li, S-H. Cho, J. Huang, B. Simonsen, S. Yang, L.M. Contreras. “Multiple small RNAs interact to co-regulate ethanol tolerance in Zymomonas mobilis” Frontiers in Bioengineering and Biotechnology, 8 (2020). Available from: doi.org/10.3389/fbioe.2020.00155.
55. Lukasiewicz, A. and L.M. Contreras . “Antisense probing of dynamic RNA structures” Methods (2020). Available from: doi.org/10.1016/j.ymeth.2020.01.015.
54. E.K. Bowman, M.K. Mihailovic, B. Li, and L.M. Contreras. “Bioinformatic Application of Fluorescence-based in vivo RNA Regional Accessibility Data to Identify Novel sRNA Targets” In: Arluison V., Wien F. (eds) RNA Spectroscopy. Methods in Molecular Biology, vol 2113. Humana, New York, NY (2020). Available from: doi.org/10.1007/978-1-0716-0278-2_5.
53. J.C. Gonzalez-Rivera, A.A. Orr, S.M. Engels, J.M. Jakubowski, M.W. Sherman, K.N. O’Connor, T. Matteson, B.C. Woodcock, L.M. Contreras and P. Tamamis. “Computational evolution of an RNA-binding protein towards enhanced oxidized RNA binding” Computational and Structural Biotechnology Journal, 28:137-152 (2020). Available from: doi.org/10.1016/j.csbj.2019.12.003
52. K. Haning, S.M. Engels, P. Williams, M. Arnold, and L.M. Contreras. “Applying a new REFINE approach in Zymomonas mobilis identifies novel sRNAs that confer improved stress tolerance phenotypes” Frontiers in Microbiology. (2020). Available from: doi.org/10.3389/fmicb.2019.02987
2019
51. A.M. Rojano-Nisimura , K. Haning, J. Janovsky, K.A. Vasquez, J.P. Thompson, and L.M. Contreras (2019).” Codon selection affects recruitment of ribosome-associating factors during translation”. ACS Synthetic Biology (In press). Available from: doi.org/10.1021/acssynbio.9b00344 Paper featured in issue’s cover.

50. A. Chen, M.W. Sherman, and L.M. Contreras. “Discovery and Characterization of Native Deinococcus radiodurans Promoters for Tunable Gene Expression”. Applied and Environmental Microbiology, AEM.01356-19 (2019). Available from: doi.org/10.1128/aem.01356-19 Paper featured in issue’s cover.

49. A. N. Leistra, N.C. Curtis and L.M. Contreras. “Regulatory Non-coding sRNAs in Bacterial Metabolic Pathway Engineering ”Metabolic Engineering, 52:190-214 (2019). Available from: doi.org/10.1016/j.ymben.2018.11.013
2018
48. A. Chen, B. K. Keitz, L.M. Contreras. “Biological links between nanoparticle biosynthesis and stress responses in bacteria” Mexican Journal of Biotechnology, 3(4):44-69 (2018). Available from: doi.org/10.29267/mxjb.2018.3.4.44
47. Y. Li, K. Reyes, J. Vazquez-Anderson, Y. Wang, L.M. Contreras , and W. Powell. “A knowledge gradient policy for sequencing experiments to identify the structure of RNA molecules using a sparse additive believe model.” INFORMS Journal on Computing, 30(4) (2018). Available from: doi.org/10.1287/ijoc.2017.0803
46. M. K. Mihailovic, J. Vazquez-Anderson, Y. Li, P. Vimalathas, V. Fry, R.A. Lease, W. Powell, and L.M. Contreras. “High-throughput in vivo mapping of RNA accessible interfaces to identify functional sRNA binding sites.” Nature Communications, 9:4084 (2018)Available from: doi.org/10.1038/s41467-018-06207-z.
45. J. K. Villa, Y. Su, L.M. Contreras and M. Hammond. “Synthetic Biology of Small RNAs and Riboswitches”, Book chapter, in Papenfort, K, Storz, G (eds.), Regulating with RNA in Bacteria and Archaea, 6(3) (2018). ASM. Available from: doi.org/10.1128/microbiolspec.RWR-0007-2017.
44. L.M. Contreras . “Methods and advances in RNA characterization and design” Methods, 143:1-3 (2018). Editorial Article; invited issue editor. Available from: doi.org/10.1016/j.ymeth.2018.06.003.

43. A. N. Leistra, G. Gelderman, S. W. Sowa, A. Moon-Walker, H. M. Salis, and L.M. Contreras. “A Canonical Biophysical Model of the CsrA Global Regulator Suggests Flexible Regulator-Target Interactions” Scientific Reports, 8:9892 (2018). Available from: doi.org/10.1038/s41598-018-27474-2.
42. K. Baldridge, M. Jora, A. Maranhao, M. Quick, B. Addepalli, J. Brodbelt, A. Ellington, P. Limbach and L.M. Contreras. “Directed Evolution of Heterologous tRNAs Leads to Reduced Dependence on Post-transcriptional Modifications” ACS Synthetic Biology, 7(5):1315-1327 (2018). Available from: doi.org/10.1021/acssynbio.7b00421.
41. X. Wang, Q He, Y. Yang, J. Wang, K. Hanning, Y. Hu, B. Wu, M. He, Y. Zhang, J. Bao, L.M. Contreras, and S. Yang “Advances and Prospects in Metabolic Engineering of Zymomonas mobilis,” Metabolic Engineering, S1096-7176(18)30036-3 (2018). Available from: doi.org/10.1016/j.ymben.2018.04.001.
40. M. Sherman and L.M. Contreras “Computational approaches in design of nucleic acid-based therapeutics” Current Opinion in Biotechnology, 18(53):232-239 (2018). Available from: doi.org/10.1016/j.copbio.2017.12.001.
39. A. Leistra, M.K Mihailovic and L.M. Contreras “Fluorescence-based Methods for Characterizing RNA interactions in vivo,” in: Arluison V., Valverde C. (eds) Bacterial Regulatory RNA. Methods in Molecular Biology, vol 1737. Humana Press, New York, NY. Available from: doi.org/10.1007/978-1-4939-7634-8_9.
38. A.A. Orr, J.C. Gonzalez-Rivera, M. Wilson, P.R. Bhikha, D. Wang, L.M. Contreras, and P. Tamamis. “A high-throughput and rapid computational method for screening of RNA post-transcriptional modifications that can be recognized by target proteins,” Methods, 143:34-47 (2018). Available from: doi.org/10.1016/j.ymeth.2018.01.015.
2017
37. S-H. Cho, K. Haning, W. Shen, C. Blome, R. Li, S. Yang, and L.M. Contreras, “Identification and characterization of 5′ untranslated regions (5’UTRs) in Zymomonas mobilis as regulatory biological parts,” Frontiers in Microbiology, section Microbial Physiology and Metabolism, 8:2432 (2017). Available from: doi:10.3389/fmicb.2017.02432.
36. A. Chen, L.M. Contreras, and Benjamin Keitz, “Imposed Environmental Stresses Facilitate Cell-free Nanoparticle Formation by Deinococcus radiodurans,” Applied and Environmental Microbiology, 83 (18): e00798-17 (2017). Available from: doi:10.1016/10.1128/AEM.00798-17
35. J. K. Villa, P. Amador, J. Janovsky, A. Bhuyan, R. Saldanha, T. J. Lamkin, and L.M. Contreras, “Genome-wide search for ionizing radiation responsive 5’UTRs in Deinococcus radiodurans reveals post-transcriptional regulation in the Radiation and Desiccation Response (RDR) for DNA gyrase subunit A,” Applied and Environmental Microbiology, 83(12): e00039-17 (2017). Available from: doi:10.1128/AEM.00039-17
34. A. Leistra, P. Amador, A. Buvanendiran, A. Moon-Walker andL.M. Contreras “Rational modular RNA engineering based on in vivoprofiling of structural accessibility” ACS Synthetic Biology (2017). Available from: doi:10.1016/10.1021/acssynbio.7b00185
33. J. Vazquez-Anderson, M. K. Mihailovic, K.C. Baldridge, K. Reyes, K. Haning, S. H. Cho, P. Amador, W. Powell, and L.M. Contreras, “Optimization of a novel biophysical model using large scale in vivo antisense hybridization data displays improved prediction capabilities of structurally accessible RNA regions,” Nucleic Acids Research Journal, 45(9):5523–5538 (2017). Available from: doi:10.1093/nar/gkx115
32. M.K. Mihailovic, A. Chen, J.C. Gonzalez-Rivera, and L.M. Contreras, “Defective RNPs, mistakes in RNA processing and Diseases,” Biochemistry, 56(10):1367–1382 (2017). (Invited Article). Available from: doi:10.1016/10.1021/acs.biochem.6b01134
31. S. Sowa, G. Gelderman, A. N. Leistra, A. Buvanendiran, S. Lipp, A. Pitaktong, T. Romeo, M. Baldea, and L.M. Contreras, “Integrative FourDomics approach profiles the target network of the carbon storage regulatory system,” Nucleic Acids Research Journal, 45(4): 1673-1686 (2017). Available from: doi:10.1093/nar/gkx048.
2016
30. S. Yang, Q. Fei, Y. Zhang, L.M. Contreras , S. M. Utturkar, S. D. Brown, M. E. Himmel and M. Zhang. “Zymomonas mobilis as a Model System for Production of Biofuels and Biochemicals,” Microbial Biotechnology, 9(6): 699–717 (2016). Available from: doi:10.1111/1751-7915.12408.
29. K. Vasquez, T. Hatridge, N. Curtis, and L.M. Contreras, “Modifying translation timing between protein domains by regulating affinity between mRNAs and the ribosomal anti-Shine-Dalgarno sequence results in predictable solubility changes,” ACS Synthetic Biology, 5(2): 133-145 (2016). Available from: doi:10.1021/acssynbio.5b00193. Paper featured in issue’s cover.

2015
27. S. Sowa, G. Gelderman, and L.M. Contreras, “Advances in synthetic dynamic circuits: using novel synthetic parts to engineer new generations of gene oscillations,” Current Opinion in Biotechnology, 36:161-167 (2015). (Invited Article). Available from: doi:10.1016/j.copbio.2015.08.020.
26. K. Haning, S-H. Cho, and L. M. Contreras, “Strain Engineering via regulatory noncoding RNA mechanisms: not a one-blueprint fits all,” Current Opinion in Chemical Engineering, 10:25-34 (2015). (Invited Article). Available from: doi:10.1016/j.coche.2015.07.008
25. C-H. Tsai, R. Liao, B. Chou, and L.M. Contreras, “Transcriptional Analysis of Deinococcus radiodurans reveals small RNAs that are differentially expressed under ionizing radiation,” Applied and Environmental Microbiology, 81(5): 1745-55 (2015). Available from: doi:10.1128/AEM.03709-14
24. G. Gelderman, A. Sivakumar, S. Lipp, L.M. Contreras, “Adaptation of Tri-molecular fluorescent complementation allows assaying of regulatory Csr RNA-protein interactions in bacteria,” Biotechnology and bioengineering, 112(2): 365-375 (2015). Available from: doi:10.1002/bit.25351.
23. K. Baldridge, J. Zavala, J. Surratt, K. Sexton, and L.M. Contreras, “Cellular RNA is chemically modified by exposure to air pollution mixtures,” Inhalation Toxicology, 27(1):74-82 (2015). Available from: doi:10.3109/08958378.2014.987361.
22. C-H. Tsai, R. Liao, B. Chou, M. Palumbo, and L.M. Contreras, “Genome-wide analysis in bacteria shows sRNA enrichment in long and conserved intergenic regions,” Journal of Bacteriology, 197(1):40-50 (2015). Available from: doi:10.1128/JB.02359-14.
21. S. Sowa, J. Vazquez-Anderson, C. Clark, R. De La Peña, K. Dunn, E. Fung, M. Khoury, and L.M.Contreras,“Exploiting post-transcriptional regulation to probe RNA structures in vivo via fluorescence,” Nucleic Acids Research, 43(2):e13 (2015). Available from: doi:10.1093/nar/gku1191
2014
20. K. Haning, S-H. Cho, and L.M.Contreras, ”Small RNAs in Mycobacteria: an unfolding story,” Frontiers in cellular infection and microbiology, 4(96):1-11 (2014). (Invited Article). Available from: doi:10.3389/fcimb.2014.00096
19. S. Sowa, M. Baldea, and L.M.Contreras, “Optimizing metabolite production using periodic oscillations,” PLOS Computational Biology, 10(6): e1003658 (2014). Available from: doi:10.1371/journal.pcbi.1003658
18. S-H. Cho, R. Lei, and L.M. Contreras, “Discovery of ethanol responsive small RNAs in Zymomonas mobilis,” Applied and Environmental Microbiology, 80(14):4189-98 (2014). Available from: doi:10.1128/AEM.00429-14
17. K.Gupta, L.M.Contreras, T. Huang, L. Spruce, S. Seeholzer, M. Belfort, and G. Van Duyne,, “Quaternary arrangement of an active, native group II intron ribonucleoprotein complex revealed by small-angle X-ray scattering,” Nucleic Acids Research, 42(8): 5347-60 (2014). Available from: doi:10.1093/nar/gku140
16. K. Baldridge, and L.M.Contreras, “Functional implications of rRNA methylations in response to environmental stress,” Critical Reviews in Biochemistry and Molecular Biology, 49(1):69-89 (2014). Available from: doi:10.3109/10409238.2013.859229
2013
15. J. Vazquez-Anderson, and L.M. Contreras, “Regulatory RNAs: Charming Gene Management Styles for Synthetic Biology Applications” RNA Biology, 10(12):1778-97 (2013). Available from: doi:10.4161/rna.27102
14. C-H. Tsai, C. Baranowski, J. Livny, K. McDonough, J. Wade, and L.M. Contreras, “Identification of novel sRNAs in Mycobacterial species”, Plos One 4;8(11):e79411 (2013). Available from: doi:10.1371/journal.pone.0079411
13. L.M. Contreras, T.Huang, C.L. Piazza, D. Smith, G. Qu, G. Gelderman, J. Potratz, R. Russell, M. Belfort, “Group II intron ribosome association protects intron RNA from degradation,” RNA 19 (11): 1497–1509 (2013). Available from: doi:10.1261/rna.039073.113
12. G.Gelderman, and L.M. Contreras, “Discovery of posttranscriptional regulatory RNAs using next generation sequencing technologies,” Methods in Molecular Biology, 985:269-95 (2013). Available from: doi:10.1007/978-1-62703-299-5_14
11. S. Sowa, J. Vazquez-Anderson, and L.M. Contreras, “Capturing cellular regulation in silico using big data: a frontier for systems biology,” Current Synthetic and Systems Biology, 19;1(1):1-4 (2013). Available from: doi:10.4172/2332-0737.1000107
2012
10. L.M. Contreras, J. Kostecki, and M.P. DeLisa, “The ribosomal exit tunnel as a target for optimizing protein expression in Escherichia coli,” Journal of Biotechnology, 7 (3): 354-60 (2012). Available from: doi:10.1002/biot.201100198
Prior to 2012
9. T. Huang, T. Shaikh, K. Gupta, L.M. Contreras, R. Grassucci, G.D.Van Duyne, J. Frank, and M. Belfort, “The group II intron ribonucleoprotein precursor is a large, loosely packed structure,” Nucleic Acids Research, 39(7) : 2845-2854 (2011). Available from: doi: 10.1093/nar/gkq1202
8. W-Y. Wu, A. R.. Gillies, J.F. Hsii, L.M. Contreras, S. Oak, M.B. Perl, and D.W. Wood, “Self-cleaving purification tags re-engineered for rapid Topo® cloning,” Biotechnology Progress, 26(5):1205-12 (2010). Available from: doi:10.1002/btpr.430
7. J.M. Dichiara, L.M. Contreras, D. Smith, and M. Belfort, “Multiple small RNAs identified in Mycobacterium bovis BCG are also expressed in Mycobacterium tuberculosis and Mycobacterium smegmatis,” Nucleic Acids Research, 38 (12): 4068-4078 (2010). Available from: doi:10.1093/nar/gkq101
6. E.E. Borrero, L.M. Contreras, M.P. DeLisa, “Kinetics and reaction coordinates of the reassembly of protein fragments via forward flux sampling,” BioPhysical Journal, 98(9):1911-20 (2010). Available from: doi:10.1016/j.bpj.2009.12.4329
5. L.M. Contreras, E.E. Borrero, F. Escobedo, and M.P. DeLisa. “In silico protein fragmentation reveals the importance of critical nuclei in domain reassembly,” BioPhysical Journal, 94(5): 1575-88 (2007). Available from: doi:10.1529/biophysj.107.119651
4. L.M. Contreras-Martinez, M.P DeLisa. “Expression engineering of synthetic antibodies using ribosome display”. In: M. Dyson, Y. Durocher, editors. Expression Systems: Methods Express. 1st ed. Oxfordshire, UK: Scion Publishing; pp. 29-52 (2007).
3. L.M. Contreras and M.P. DeLisa, “Intracellular ribosome display via SecM translation arrest as a selection for antibodies with enhanced cytosolic stability,” Journal of Molecular Biology, 372(2): 513-524 (2007). Available from: doi:10.1016/j.jmb.2007.06.070
2. L.M. Contreras, J. Walls J. “Feedback from workshop participants”. In: R. Gray, S. Hemami, E. Riskin, R. Ward, S. Brainard, P. Cosman, N. Fortenberry, J. Rutledge, T. Whitney, editors. Mentoring for Engineering Academia II. ; pp. 101–124 (2007).
1. L.M. Contreras, F.Martinez-Veracoechea, P. Pohkarel, A.D. Stroock, F. Escobedo and M.P. DeLisa, “Protein translocation through a tunnel induces changes in folding kinetics: a lattice model study,”Biotechnology and Bioengineering, 94: 105-117 (2006). Available from: doi:10.1002/bit.20832
PATENTS
2. DeLisa MP, Contreras LM. “Protein Discovery Using Intracellular Ribosome Display” (Provisional Patent Application filed on June 21, 2007). Licensed by Vybion, Inc.
1. Wood D, HsII J, Oak S, Contreras L, Chestnut J. “Self-Cleaving Affinity Tags and Methods of Use”. (PCT application filed with US Patent Office February 27, 2005).

