
General Themes and Goals of our Research Group:
The general theme of our lab is discovery and characterization of RNA regulators for uncovering complex cellular phenotypes. A major application of our work is the manipulation of RNA regulatory systems to engineer more robust cellular phenotypes that can better withstand natural environmental or chemical stresses (like those imposed by the incorporation of heterologous metabolic pathways). Some of the critical tools used in our lab include biochemical approaches, systems biology approaches, genetic studies, large-scale “omics” data, molecular engineering, synthetic biology and bioinformatics. We also collaborate with a number of top scientists to incorporate structural biology, molecular dynamics modeling, kinetic simulations and other types of approaches into our work.
Areas of Research:
Characterization of non-coding RNA regulatory networks
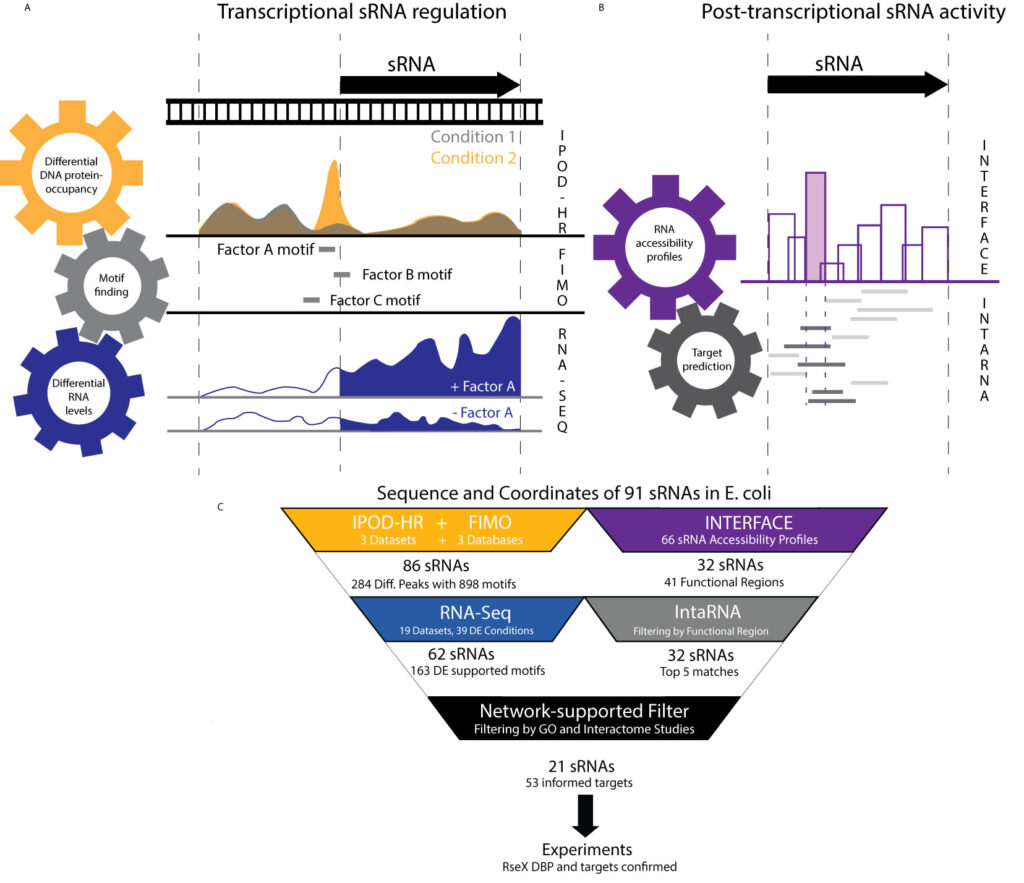
It is becoming clear that RNAs function in the context of highly intricate and interrelated networks. In our lab, we have developed a number of tools to characterize the molecular details and diversity of these interactions as well as their ultimate effects in regulating the versatility of RNA functioning.
RNA conformational changes in vivo and the relevance of these structural features for gene targeting approaches

RNAs highly depend on their conformational flexibility for stability and function. One of our interests is to understand specific mechanisms by which RNAs are stabilized in vivo by other biological molecules and to apply this knowledge to the engineering of a wide range of RNAs with robust and controllable properties.
Engineering synthetic RNA elements
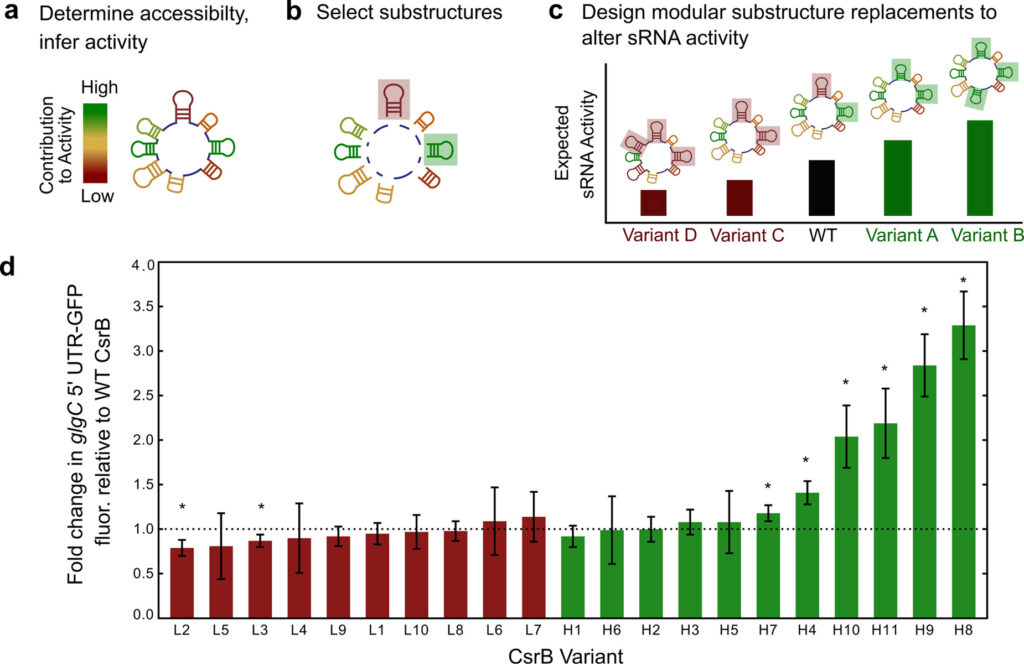
This area focuses on engineering new RNA-based elements with powerful roles in metabolic regulation. We are especially interested in elucidating ways to integrate the design of these individual molecules within the global architecture of cellular metabolism.
Characterizing RNA-mediated regulation pathways that are affected by various sources of oxidative stresses to build early disease diagnostics
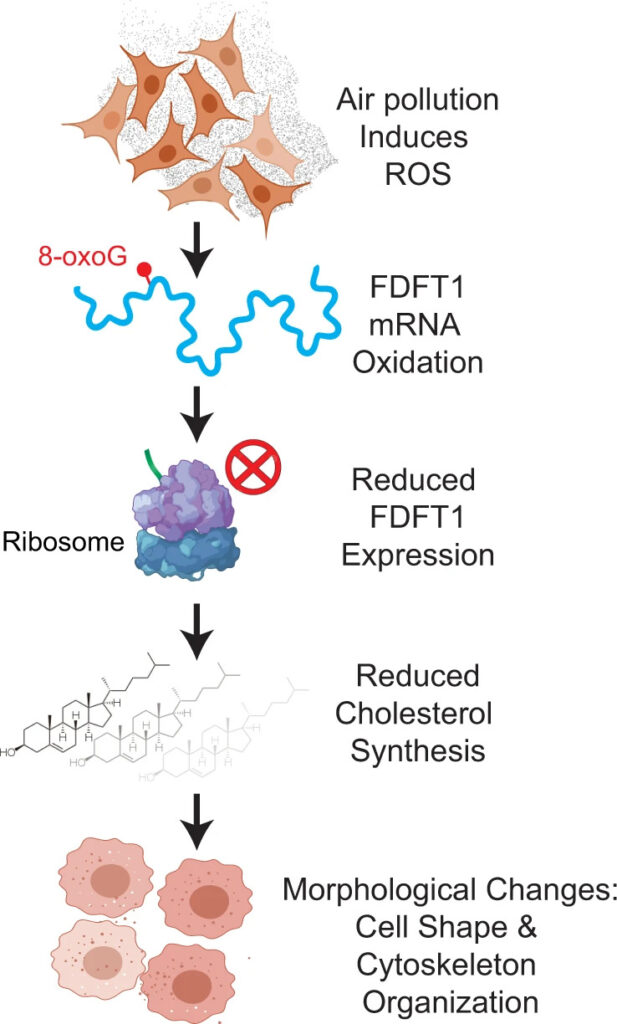
It is well understood that RNAs control a variety of essential cellular pathways and responses. On the other hand, oxidative stresses have been outlined as a major cause of aging and diseases. This area of our research focuses on understanding the molecular attributes that lead to altered mechanisms of RNA regulation within cells in response to changing environmental inputs that are encountered on a daily basis. Some of the stressors that we are studying include radiation and various air pollutants. A major interest in this area is global cellular epitranscriptomics changes that are induced by oxidative stresses.
Additional Resources
Get to know Dr. Contreras and the lab!
“Using synthetic biology and machine learning tools to characterize and engineer RNAs” (2020)
Why Where You Live Can Impact Lung Health (2020)
“In vivo Methods to Characterize and Design Regulatory sRNAs in Bacteria” (2018)
“Noncoding regulatory RNAs and oxidative stress responses” (2018)“Better living through Microbes” (2015)
*Research overview and portions of figures created using BioRender.com