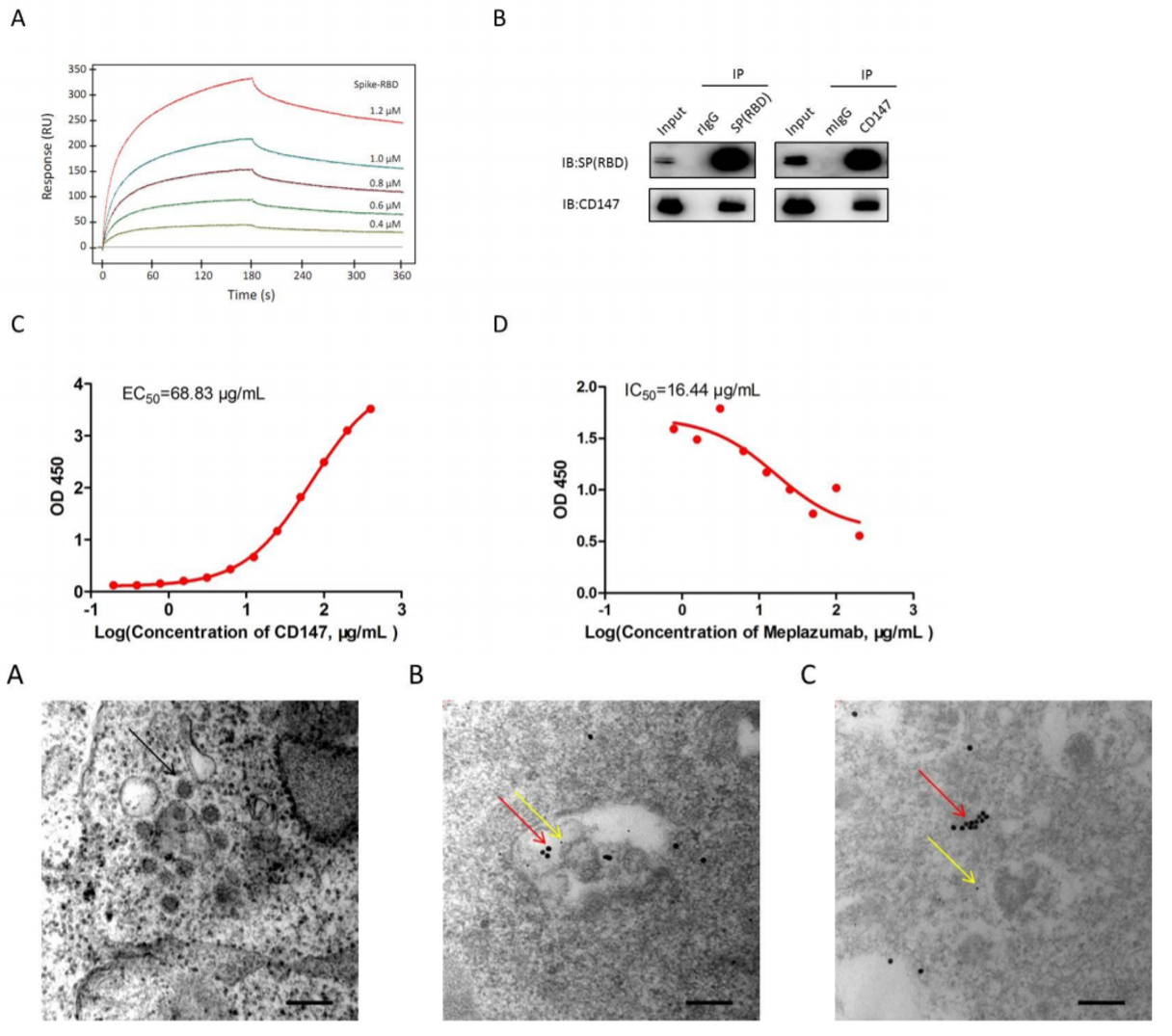
ACE2 is well established as the primary target for viral attachment and entry for both SARS-CoV-1 and SARS-CoV-2 (Li et al., 2005; Zhang et al., 2020). In addition, several other molecules have been discovered to play an accessory role in permitting viral entry into host cells. One of these accessory proteins is a molecule called TMPRSS2, a serine protease that acts to prime the viral spike (S) protein of SARS-CoV-1 prior to interaction with ACE2 and was also found to be implicated in the attachment of SARS-CoV-2 (Hoffman et al., 2020). More recently, CD147 was found to be a novel mediator of viral attachment in SARS-CoV-2 infection. CD147 is a membrane-spanning glycoprotein that is prominently associated with tumor development, protection from plasmodium invasion, and viral infection: it has been shown to inhibit signaling pathways of the tumor suppressor p53, block red blood cell infection of the protozoan that causes malaria P. falciparum, and facilitate HIV invasion (Huang et al., 2014; Zhang et al., 2017; Pushkarsky et al., 2001). Previously, Wang et al. showed that CD147 facilitates SARS-CoV-1 entry, which could be inhibited by CD147-antagonist peptide-9 in the HEK293T cell line (Chen et al., 2005). Here, they first demonstrated that humanized anti-CD147 monoclonal antibody Meplazumab competitively inhibited binding of the S protein and CD147 – compared to controls, CD147-blocked cells displayed significantly less infection by SARS-CoV-2. Additionally, Surface Plasmon Resonance (SPR, Top figure 1A) and co-immunoprecipitation with anti-CD147 and anti-S protein (Top figure 1B) confirmed the interaction of the two molecules with micromolar affinity (Kd = 1.85 x 10-7 M); and competitive direct ELISA (Top figure 1C-D) confirmed a dose-dependent interaction and its inhibition by Meplazumab. Follow-up observation by electron microscopy in infected Vero E6 cells (Bottom figure 1A-C) demonstrated significant co-localization of CD147 and S protein in viral inclusion bodies (Wang et al., 2020). The discovery of novel routes of entry provides unique opportunities for supplementing the molecular arsenal against SARS-CoV-2.
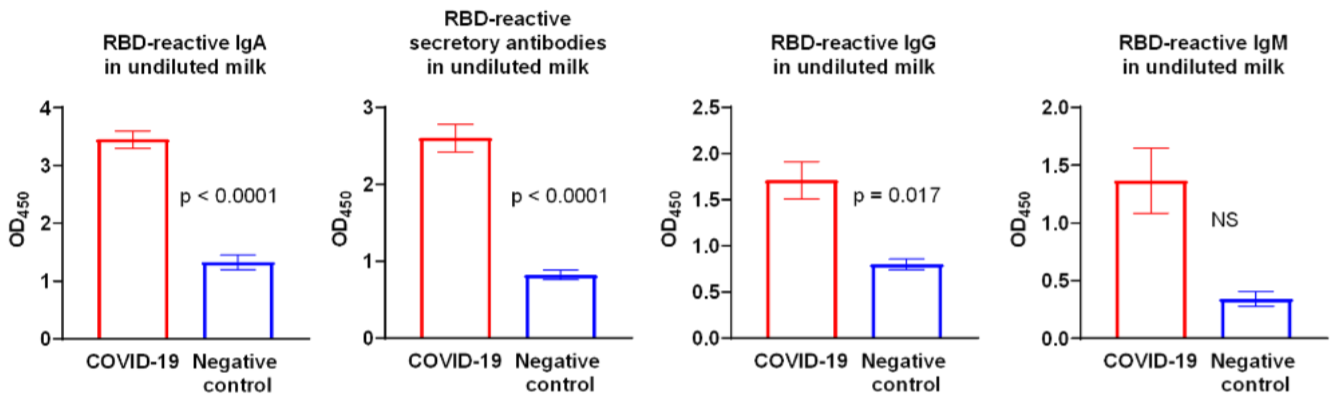
Convalescent plasma screening for IgG and IgM against pathogens is an important method for [relatively] quick identification of potentially neutralizing epitopes on those pathogens. However, of growing import is the transmission of the potentially protective IgA from mothers previously infected with SARS-CoV-2 to their babies through breastfeeding. In a recent study, fifteen milk samples were obtained from previously infected donors and compared to post-outbreak controls (the post-infection samples were obtained between 14 and 30 days after symptoms abated in these patients). In twelve of the fifteen samples, purified IgA demonstrated significant reactivity to the receptor-binding domain (RBD) of the S protein of SARS-CoV-2. Overall, the optical density measurements for the COVID-19-recovered patients were significantly greater than control measurements for IgA, IgG and secretory-type antibodies specific for the RBD (Fox et al., 2020). Growing support for the intestine’s role in proper immune system development and functioning has lent credence to a connection between this environment and that of the IgA-protected mucosa of the mammary glands – the so-called “entero-mammary link”. The intestines, being a mucosal environment, are predominant sites of IgA production and secretion by intraepithelial B cells proliferating in mesenteric lymph nodes. The authors postulate that IgA specific for SARS-CoV-2 may have originated in the gut, as previous mammary IgA has been found to exhibit specificity for enteric antigens (Seppo et al., 2017). This work suggests an important role for IgA in the immune response against SARS-CoV-2.
Written by: Parker Davis
Edited by: Jina Zhou and Esther Melamed
5/18/2020
References
Chen, Z., Mi, L., Xu, J., Yu, J., Wang, X., Jiang, J., Xing, J., Shang, P., Qian, A., Li, Y., Shaw, P. X., Wang, J., Duan, S., Ding, J., Fan, C., Zhang, Y., Yang, Y., Yu, X., Feng, Q., … Zhu, P. (2005). Function of HAb18G/CD147 in Invasion of Host Cells by Severe Acute Respiratory Syndrome Coronavirus. The Journal of Infectious Diseases, 191(5), 755–760. https://doi.org/10.1086/427811
Fox, A., Marino, J., Amanat, F., Krammer, F., Hahn-Holbrook, J., Zolla-Pazner, S., & Powell, R. L. (n.d.). Evidence of a significant secretory-IgA-dominant SARS-CoV-2 immune response in human milk following recovery from COVID-19. https://doi.org/10.1101/2020.05.04.20089995
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., Schiergens, T. S., Herrler, G., Wu, N. H., Nitsche, A., Müller, M. A., Drosten, C., & Pöhlmann, S. (2020). SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell, 181(2), 271-280.e8. https://doi.org/10.1016/j.cell.2020.02.052
Huang, Q., Li, J., Xing, J., Li, W., Li, H., Ke, X., Zhang, J., Ren, T., Shang, Y., Yang, H., Jiang, J., & Chen, Z. (2014). CD147 promotes reprogramming of glucose metabolism and cell proliferation in HCC cells by inhibiting the p53-dependent signaling pathway. Journal of Hepatology, 61(4), 859–866. https://doi.org/10.1016/j.jhep.2014.04.035
Li, W., Zhang, C., Sui, J., Kuhn, J. H., Moore, M. J., Luo, S., Wong, S. K., Huang, I. C., Xu, K., Vasilieva, N., Murakami, A., He, Y., Marasco, W. A., Guan, Y., Choe, H., & Farzan, M. (2005). Receptor and viral deLi, W., Zhang, C., Sui, J., Kuhn, J. H., Moore, M. J., Luo, S., Wong, S. K., Huang, I. C., Xu, K., Vasilieva, N., Murakami, A., He, Y., Marasco, W. A., Guan, Y., Choe, H., & Farzan, M. (2005). Receptor and viral determinants of SARS-c. EMBO Journal, 24(8), 1634–1643. https://doi.org/10.1038/sj.emboj.7600640
Pushkarsky, T., Zybarth, G., Dubrovsky, L., Yurchenko, V., Tang, H., Guo, H., Toole, B., Sherry, B., & Bukrinsky, M. (2001). CD147 facilitates HIV-1 infection by interacting with virus-associated cyclophilin A. Proceedings of the National Academy of Sciences of the United States of America, 98(11), 6360–6365. https://doi.org/10.1073/pnas.111583198
Seppo, A. E., Savilahti, E. M., Berin, M. C., Sampson, H. A., & Järvinen, K. M. (2017). Breast milk IgA to foods has different epitope specificity than serum IgA—Evidence for entero-mammary link for food-specific IgA? Clinical and Experimental Allergy, 47(10), 1275–1284. https://doi.org/10.1111/cea.12945
Wang, K., Chen, W., Zhou, Y.-S., Lian, J.-Q., Zhang, Z., Du, P., Gong, L., Zhang, Y., Cui, H.-Y., Geng, J.-J., Wang, B., Sun, X.-X., Wang, C.-F., Yang, X., Lin, P., Deng, Y.-Q., Wei, D., Yang, X.-M., Zhu, Y.-M., … Chen, Z.-N. (2020). SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. BioRxiv, 2020.03.14.988345. https://doi.org/10.1101/2020.03.14.988345
Zhang, H., Penninger, J. M., Li, Y., Zhong, N., & Slutsky, A. S. (2020). Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Medicine, 46(4), 586–590. https://doi.org/10.1007/s00134-020-05985-9
Zhang, M. Y., Zhang, Y., Wu, X. D., Zhang, K., Lin, P., Bian, H. J., Qin, M. M., Huang, W., Wei, D., Zhang, Z., Wu, J., Chen, R., Feng, F., Wang, B., Nan, G., Zhu, P., & Chen, Z. N. (2018). Disrupting CD147-RAP2 interaction abrogates erythrocyte invasion by Plasmodium falciparum. Blood, 131(10), 1111–1121. https://doi.org/10.1182/blood-2017-08-802918
Leave a Reply