The role of interferons in fending against viruses has been known and investigated for decades, and in light of this, interferon therapy is well-established for treating viral infections and even some cancers (Sen, 2001; Samuel, 2001; Korenman et al., 1991; Tanner et al., 1990). Interferon therapy has recently been suggested as an option for use in COVID-19 treatment to mitigate the cytokine storm symptoms observed in early infection (Sallard et al., 2020; Prompetchara, Ketloy, Palaga, 2020). Less is known, however, about the exact molecular mechanisms that occur downstream due to interferon release.
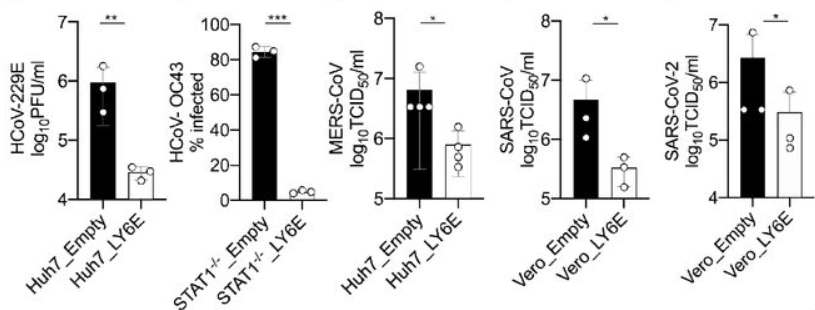
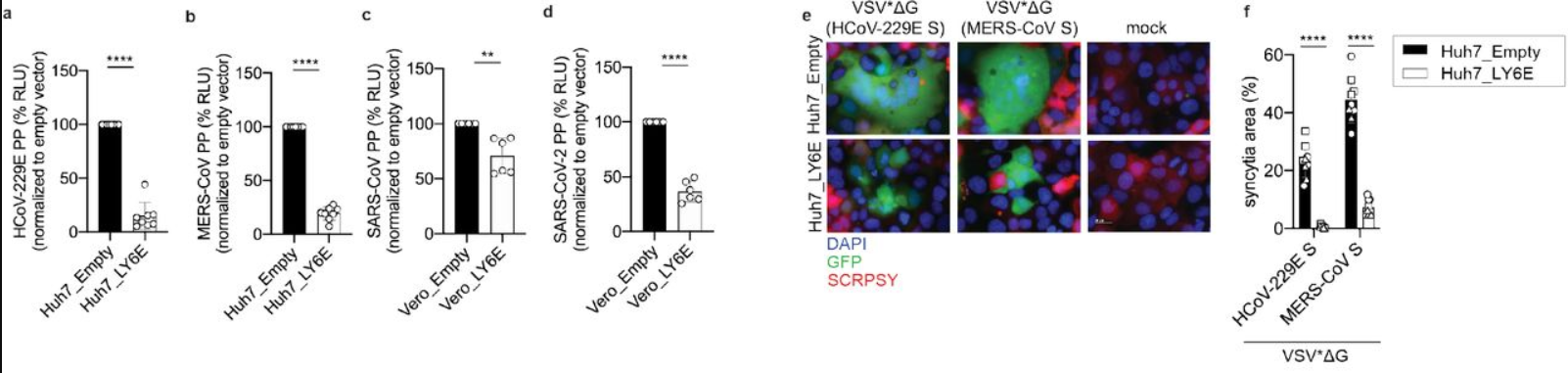
Lymphocyte antigen 6 complex, locus E (LY6E) is one such molecule involved in the downstream action of interferon release. It is one protein in a class known as Interferon-Stimulated Genes (ISGs) and plays a central role in various immune and tumorigenic pathways (Xu et al., 2014; Yeom et al., 2016; Alhossiny et al., 2016). Pfaender et al. developed a study to determine the involvement of LY6E in SARS-CoV-2 infection. They screened over 350 previously known ISG DNA sequences for ability to inhibit the human coronavirus strain HCoV-229E; LY6E stood out as a potent inhibitor of infection. LY6E also significantly inhibited infection by HCoV-OC43, MERS-CoV, SARS-CoV, and SARS-CoV-2, in addition to hepatitis virus and murine coronavirus (Figure 1). To determine the effect that endogenous LY6E has in controlling infection, the CRISPR-Cas9 system was used to ablate LY6E in human lung adenocarcinoma cells, which exhibited significant HCoV-229E infection compared to un-ablated control cells. Subsequent reconstitution with CRISPR-resistant LY6E restored the antiviral activity in the knockout cells. Several mechanisms of antiviral action were tested for LY6E: it did not exhibit a capacity to restrict HCoV-229E attachment to host cells, nor did it demonstrate any effect on the expression of CD13, the host cell surface receptor for HCoV-229E. In a syncytia formation assay, however, vesicular stomatitis virus (VSV) pseudoparticles were made to express spike (S) proteins from each of the MERS, SARS, and SARS-2 coronaviruses and LY6E significantly inhibited syncytia formation by blocking membrane fusion (Pfaender et al., 2020). These data underscore the effect of interferon therapies for preventing viral infection. Future work should focus on elucidating ISG pathways, which may be key in developing more effective therapies, tailored to a given disease.
Viruses are not technically living beings and are characterized as “obligate intracellular” pathogens, meaning that they rely on their host cell’s machinery to replicate and propagate infections. It follows, then, that virus- and host-derived molecules mingle inside the infected cell, sometimes with the effect of modulating the activity of the molecule with which they interact. Liang et al. analyzed the network between SARS-CoV-2 and HEK293 cell proteins and developed a map of 251 animal (HEK) proteins that coordinate with the ~30 known coronavirus proteins to produce upwards of 630 high-confidence interactions (Figure 3). The authors noted several specific protein-protein synergies that they hypothesized to be involved in SARS-CoV-2 pathogenesis: between animal C1QBP and CoV nonstructural proteins [nsp] 9 and 10; between animal RAE1 and CoV open reading frame [ORF] 6; and between animal nuclear factor-kappa B-repressing protein [NKRF] and CoV nsp9, nsp10, nsp12, nsp13, and nsp15.
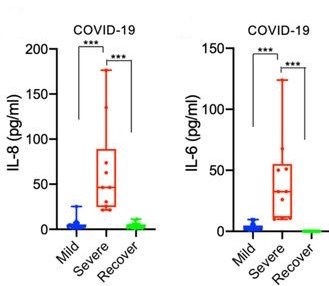
To determine which of these may be significant, peripheral blood mononuclear cells (PBMCs) from 35 COVID-19 patients were analyzed and revealed a heightened innate response compared to healthy controls, and especially elevations in interleukin (IL) 8 and IL6 that were significant in severe patients compared to mild patients and controls (Figure 4). In normal healthy humans, nuclear factor-kappa B (NF-κB) acts as a transcriptional hub to regulate the expression of many genes involved in the immune response and other biological systems (Bender et al., 2020; Meng et al., 2020; Tripathi, Aggarwal, 2006). Among these complex and interwoven pathways, it serves to induce the expression of IL8 and IL6, molecules critical to the production of inflammation in the early stages of the innate immune response (Kunsch, Rosen, 1993). NKRF is a molecule whose basic function is in the infancy of understanding, but which seems to be especially involved in inhibiting the action of NF-κB by degrading its downstream RNA products (Coccia et al., 2017; Memet et al., 2017). The elevated IL8/6 in severe COVID-19 patients and the observed interaction of NKRF with the coronavirus nonstructural proteins led the authors to hypothesize that nsp9/10/12/13/15 associate with NKRF during infection, thereby sequestering it and preventing it from inhibiting the induction of IL8 and IL6 by NF-κB.
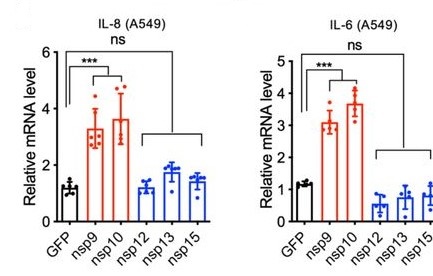
Indeed, individually expressed nsp9 and nsp10 facilitated the significant induction of IL8 and IL6 in lung epithelial cells compared to controls, whereas nsp12, nsp13, and nsp15 did not (Figure 5). In addition, degradation of NKRF by silencing RNA in vitro resulted in an induction of IL8 similar to that seen in severe COVID-19 patients, thereby providing support for the authors’ hypothesis (Liang et al., 2020).

Written by: Parker Davis
Edited by: Jina Zhou and Esther Melamed
6/2/2020
References
Alhossiny, M., Luo, L., Frazier, W. R., Steiner, N., Gusev, Y., Kallakury, B., Glasgow, E., Creswell, K., Madhavan, S., Kumar, R., & Upadhyay, G. (2016). Ly6E/K signaling to TGFβ promotes breast cancer progression, immune escape, and drug resistance. Cancer Research, 76(11), 3376–3386. https://doi.org/10.1158/0008-5472.CAN-15-2654
Bender, A. T., Tzvetkov, E., Pereira, A., Kasar, S., Przetak, M., Okitsu, S., & Vlach, J. (2020). TLR7 and TLR8 differentially activate the IRF and NF-kB pathways in specific cell types to promote inflammation. Lupus Science & Medicine, 7(Suppl 1), A74.1-A74. https://doi.org/10.1136/lupus-2020-eurolupus.138
Coccia, M., Rossi, A., Riccio, A., Trotta, E., & Santoro, M. G. (2017). Human NF-κB repressing factor acts as a stress-regulated switch for ribosomal RNA processing and nucleolar homeostasis surveillance. Proceedings of the National Academy of Sciences of the United States of America, 114(5), 1045–1050. https://doi.org/10.1073/pnas.1616112114
Korenman, J., Baker, B., Waggoner, J., Everhart, J. E., Di Bisceglie, A. M., & Hoofnagle, J. H. (1991). Long-term remission of chronic hepatitis B after alpha-interferon therapy. Annals of Internal Medicine, 114(8), 629–634. https://doi.org/10.7326/0003-4819-114-8-629
Kunsch, C., & Rosen, C. A. (1993). NF-kappa B subunit-specific regulation of the interleukin-8 promoter. Molecular and Cellular Biology, 13(10), 6137–6146. https://doi.org/10.1128/mcb.13.10.613
Liang, Q., Li, J., Guo, M., Tian, X., Liu, C., Wang, X., Yang, X., Wu, P., Xiao, Z., Qu, Y., Yin, Y., Fu, J., Zhu, Z., Liu, Z., Peng, C., & Zhu, T. (2020). Virus-host interactome and proteomic survey of PMBCs from COVID-19 patients reveal potential virulence factors influencing SARS-CoV-2 pathogenesis. BioRxiv, 2020.03.31.019216. https://doi.org/10.1101/2020.03.31.019216
Memet, I., Doebele, C., Sloan, K.E., Bohnsack, M.T. (2017). The G-patch protein NF-κB-repressing factor mediates the recruitment of the exonuclease XRN2 and activation of the RNA helicase DHX15 in human ribosome biogenesis. Nucleic Acids Research, 45(9), 5359-5374. https://doi.org/10.1101/2020.03.31.019216
Meng, Q., Liang, C., Hua, J., Zhang, B., Liu, J., Zhang, Y., Wei, M., Yu, X., Xu, J., & Shi, S. (2020). A miR-146a-5p/TRAF6/NF-kB p65 axis regulates pancreatic cancer chemoresistance: Functional validation and clinical significance. Theranostics, 10(9), 3967–3979. https://doi.org/10.7150/thno.40566
Pfaender, S., Mar, K. B., Michailidis, E., Kratzel, A., Hirt, D., V’kovski, P., Fan, W., Ebert, N., Stalder, H., Kleine-Weber, H., Hoffmann, M., Hoffmann, H. H., Saeed, M., Dijkman, R., Steinmann, E., Wight-Carter, M., Hanners, N. W., Pöhlmann, S., Gallagher, T., … Thiel, V. (2020). LY6E impairs coronavirus fusion and confers immune control of viral disease. BioRxiv, 2020.03.05.979260. https://doi.org/10.1101/2020.03.05.979260
Prompetchara, E., Ketloy, C., & Palaga, T. (2020). Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. In Asian Pacific Journal of Allergy and Immunology (Vol. 38, Issue 1, pp. 1–9). https://doi.org/10.12932/AP-200220-0772
Sallard, E., Lescure, F. X., Yazdanpanah, Y., Mentre, F., & Peiffer-Smadja, N. (2020). Type 1 interferons as a potential treatment against COVID-19. Antiviral Research, 178, 104791. https://doi.org/10.1016/j.antiviral.2020.104791
Samuel, C. E. (2001). Antiviral actions of interferons. In Clinical Microbiology Reviews (Vol. 14, Issue 4, pp. 778–809). American Society for Microbiology Journals. https://doi.org/10.1128/CMR.14.4.778-809.2001
Sen, G. C. (2001). Viruses and Interferons. Annual Review of Microbiology, 55(1), 255–281. https://doi.org/10.1146/annurev.micro.55.1.255
Tanner, D. J., Taylor, E. L., Smiles, K. A., & Peets, E. A. (1990). Intralesional interferon therapy for basal cell carcinoma. Journal of the American Academy of Dermatology, 23(4), 694–700. https://doi.org/10.1016/0190-9622(90)70276-N
Tripathi, P., & Aggarwal, A. (2006). NF-kB transcription factor: A key player in the generation of immune response. In Current Science (Vol. 90, Issue 4, pp. 519–531). www.ncbi.nlm.nih.gov/entrez/query.fcgi
Xu, X., Qiu, C., Zhu, L., Huang, J., Li, L., Fu, W., Zhang, L., Wei, J., Wang, Y., Geng, Y., Zhang, X., Qiao, W., & Xu, J. (2014). IFN-Stimulated Gene LY6E in Monocytes Regulates the CD14/TLR4 Pathway but Inadequately Restrains the Hyperactivation of Monocytes during Chronic HIV-1 Infection. The Journal of Immunology, 193(8), 4125–4136. https://doi.org/10.4049/jimmunol.1401249
Yeom, C. J., Zeng, L., Goto, Y., Morinibu, A., Zhu, Y., Shinomiya, K., Kobayashi, M., Itasaka, S., Yoshimura, M., Hur, C. G., Kakeya, H., Hammond, E. M., Hiraoka, M., & Harada, H. (2016). LY6E: A conductor of malignant tumor growth through modulation of the PTEN/PI3K/Akt/HIF-1 axis. Oncotarget, 7(40), 65837–65848. https://doi.org/10.18632/oncotarget.11670
Leave a Reply