Ultimate understanding of our options for treatment and prevention of COVID-19 will come only as a result of our ability to ascertain the functions of the various SARS-CoV-2 genes and how they interact with the host organism. I’ve written previously about some of these interactions (see updates from 6/2 and 6/15), which are involved largely with host immune regulation and viral entry. A major aspect of SARS-CoV-2 is its ability to proficiently inhibit the type I interferon response upon infection (Blanco-Melo et al., 2020). Without the production of interferon (IFN), the host lacks a critical facet of its ability to initiate a substantial antiviral state. In particular, RNA viruses like SARS-CoV-2 typically induce IFN production via a retinoic acid-inducible gene-I-like (RIG-I-like) receptor that passes the signal on to a molecular complex located on the outer mitochondrial membrane (Stetson, Medzhitov, 2006; Kawai, Akira, 2007). Previous work has demonstrated an interaction between the original SARS-CoV gene Open Reading Frame 9b (Orf9b) and the mitochondria, an interaction which was shown to significantly inhibit production of type I IFN (Shi et al., 2014). Also present in SARS-CoV-2, this gene raises speculation about a potential mechanism for IFN inhibition in COVID-19. A paper published last month in Nature Cellular & Molecular Immunology sought to explore this mechanism of interference.
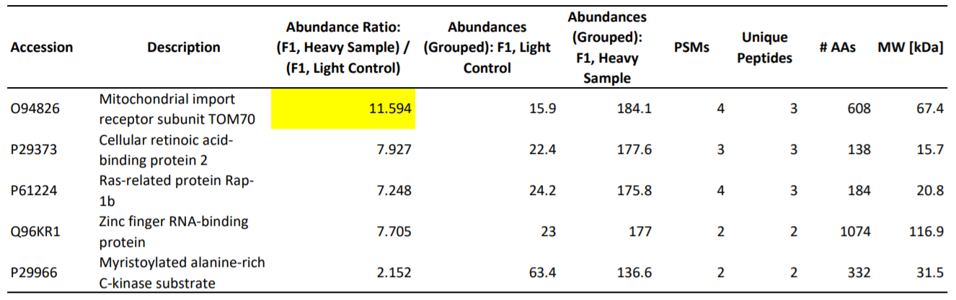
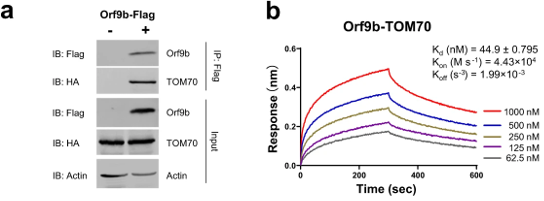
The first step in elucidating the manner of inhibition was to determine the host proteins involved in direct interactions with Orf9b. To this end, the authors utilized mass spectrometry to identify the specific proteins that make physical contact with Orf9b. Of the proteins they tested, an adapter protein called TOM70 scored the highest in its ability to interact with the viral protein (Table 1). As validation of this result, a co-immunoprecipitation assay demonstrated that TOM70 co-precipitated with Orf9b (Figure 1a). Further, biolayer interferometry yielded a binding affinity constant (Kd) of 44.9 nanomolar for the TOM70-Orf9b interaction (Figure 1b). To the authors, this suggests a strong, significant interaction between Orf9b and TOM70 in vitro.
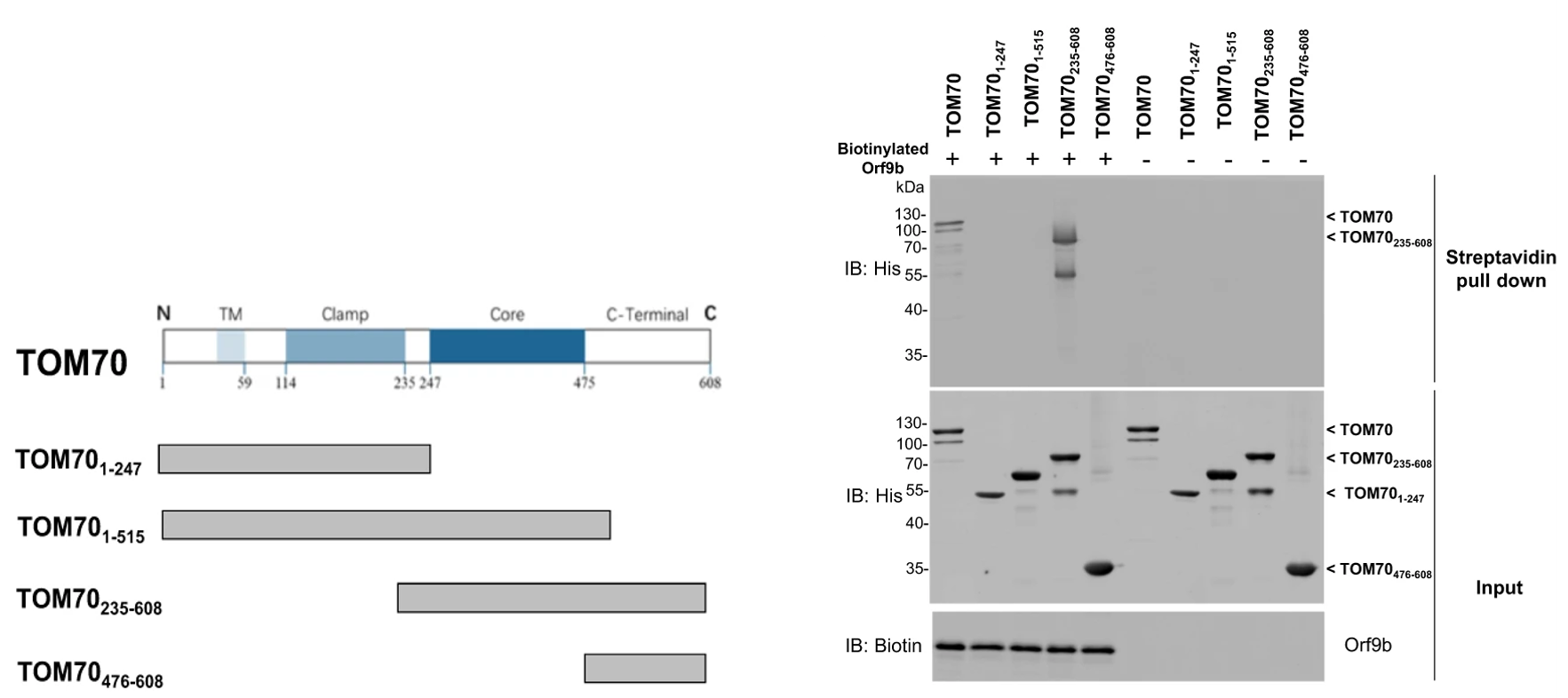
Once the protein interaction had been identified, the next step was to specify the region of TOM70 that is required for successful interaction with Orf9b. Chan et al. previously described the functions of each domain of the TOM70 gene; the authors tested each of these gene segments with Orf9b and found that a specific construct, the segment consisting of amino acids residues 235-608 co-precipitated with Orf9b (Figure 2). This segment contained the core and C-terminal domains and co-precipitated to a degree comparable to the full-length TOM70 protein, which suggests that these two domains are required for significant interaction between TOM70 and Orf9b.
As a member of the RIG-I signaling cascade, TOM70 is located in a molecular complex on the outer membrane of the mitochondria. Due to the previously demonstrated interaction with TOM70, the authors hypothesized that Orf9b may localize on the outer mitochondrial membrane in vivo. To show this, they first induced expression of Orf9b in the HEK 293T cell line. Immunostaining of these cells revealed localization of the viral protein to the outer mitochondrial membrane, as well as significant co-localization with TOM70 (Figure 3). In proteins destined for excretion or a cellular membrane, the N-terminal domain of the gene/protein codes for a signal sequence that acts as a homing device to guide the protein to its final destination. Situated on the outer mitochondrial membrane, TOM70 is one such protein. To qualitatively assess the affinity of the Orf9b-TOM70 interaction, the authors created a TOM70 construct missing its N-terminal homing domain. This would ensure that the protein ends up dislocated from its usual position in the membrane. Expression of this construct did indeed disrupt the original co-localization of Orf9b with TOM70 on the outer membrane (Figure 3). Taken together, these data support the hypothesis of a significant viral-host protein interaction.

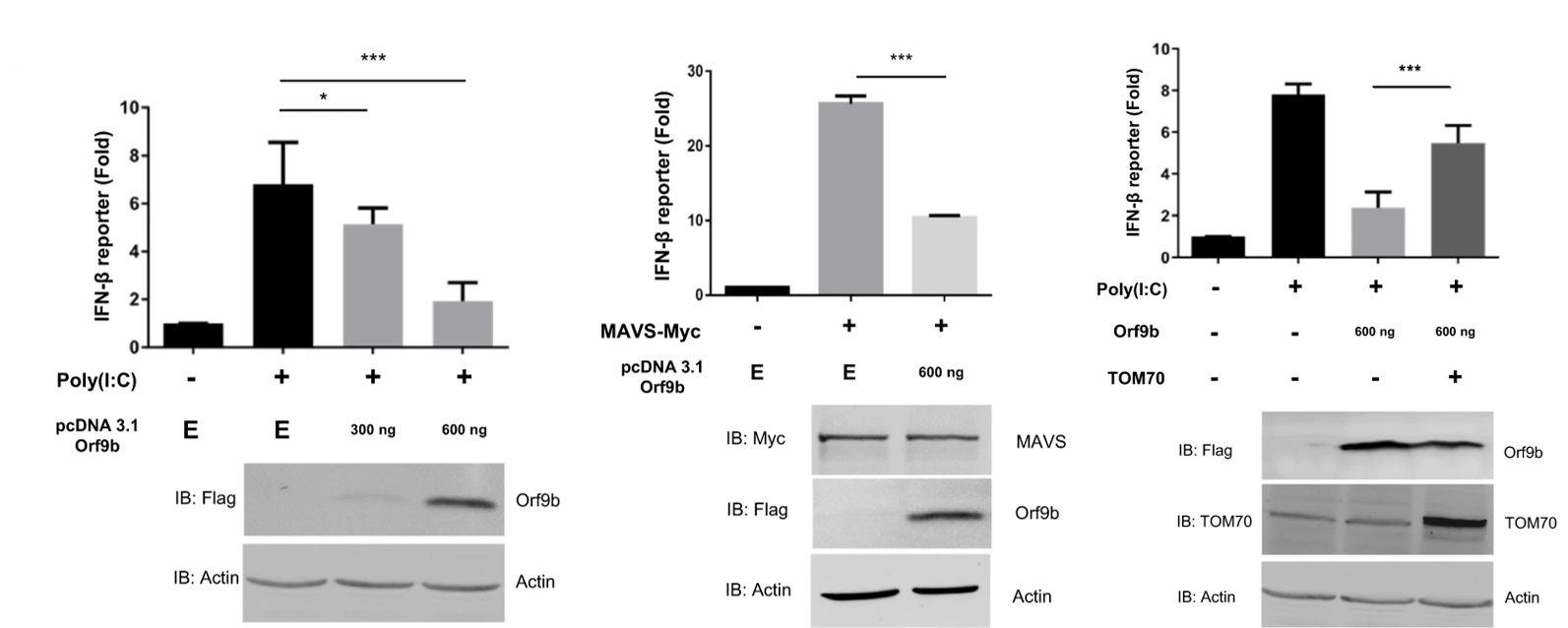
Returning to the idea of TOM70’s role in interferon production and the inhibition thereof by Orf9b, the authors observed the production of IFN-beta in either the presence or absence of Orf9b using a luciferase reporter assay. They found that the viral gene significantly reduced the activation and production of IFN-beta compared to controls (Figure 4). As a follow up, they determined that induction of TOM70 overexpression could significantly rescue type I interferon production from Orf9b inhibition. To further explore the interferon inhibition, the authors introduced TOM70 silencing RNA (siRNA) to the culture to knockout TOM70 expression in vitro. They did not observe significant suppression of interferon production upon addition of the siRNA. This indicates a potential specificity of Orf9b for TOM70 in the inhibition of interferon activation and production.
In all, this paper demonstrates the localization of Orf9b to the outer mitochondrial membrane and its suppression of type I interferon production through an interaction with the adapter protein TOM70. The authors present two possible explanations for the inhibiting action of Orf9b: 1) A separate adapter protein HSP90 normally interacts with TOM70 to induce interferon production, and Orf9b may compete with this protein for binding to TOM70, and 2) Patients with dysfunctional TOM70 tend to suffer from lactic acidosis, and interaction of Orf9b with TOM70 may induce production of lactic acid which has been shown to inhibit type I interferon production. The data presented here represent a potentially novel route for the development of therapies aimed at restoring the critical interferon response in COVID-19 patients.
References
Blanco-Melo, D., Nilsson-Payant, B. E., Liu, W. C., Uhl, S., Hoagland, D., Møller, R., Jordan, T. X., Oishi, K., Panis, M., Sachs, D., Wang, T. T., Schwartz, R. E., Lim, J. K., Albrecht, R. A., & TenOever, B. R. (2020). Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell, 181(5), 1036-1045.e9. https://doi.org/10.1016/j.cell.2020.04.026
Chan, N. C., Likić, V. A., Waller, R. F., Mulhern, T. D., & Lithgow, T. (2006). The C-terminal TPR Domain of Tom70 Defines a Family of Mitochondrial Protein Import Receptors Found only in Animals and Fungi. Journal of Molecular Biology, 358(4), 1010–1022. https://doi.org/10.1016/j.jmb.2006.02.062
Jiang, H.-W., Zhang, H.-N., Meng, Q.-F., Xie, J., Li, Y., Chen, H., Zheng, Y.-X., Wang, X.-N., Qi, H., Zhang, J., Wang, P.-H., Han, Z.-G., & Tao, S.-C. (2020). SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cellular & Molecular Immunology, 1–3. https://doi.org/10.1038/s41423-020-0514-8
Kawai, T., & Akira, S. (2007). Antiviral signaling through pattern recognition receptors. Journal of Biochemistry (Vol. 141, Issue 2, pp. 137–145). Oxford Academic. https://doi.org/10.1093/jb/mvm032
Shi, C.-S., Qi, H.-Y., Boularan, C., Huang, N.-N., Abu-Asab, M., Shelhamer, J. H., & Kehrl, J. H. (2014). SARS-Coronavirus Open Reading Frame-9b Suppresses Innate Immunity by Targeting Mitochondria and the MAVS/TRAF3/TRAF6 Signalosome. The Journal of Immunology, 193(6), 3080–3089. https://doi.org/10.4049/jimmunol.1303196
Stetson, D. B., & Medzhitov, R. (2006). Type I Interferons in Host Defense. Immunity (Vol. 25, Issue 3, pp. 373–381). Cell Press. https://doi.org/10.1016/j.immuni.2006.08.007
Leave a Reply