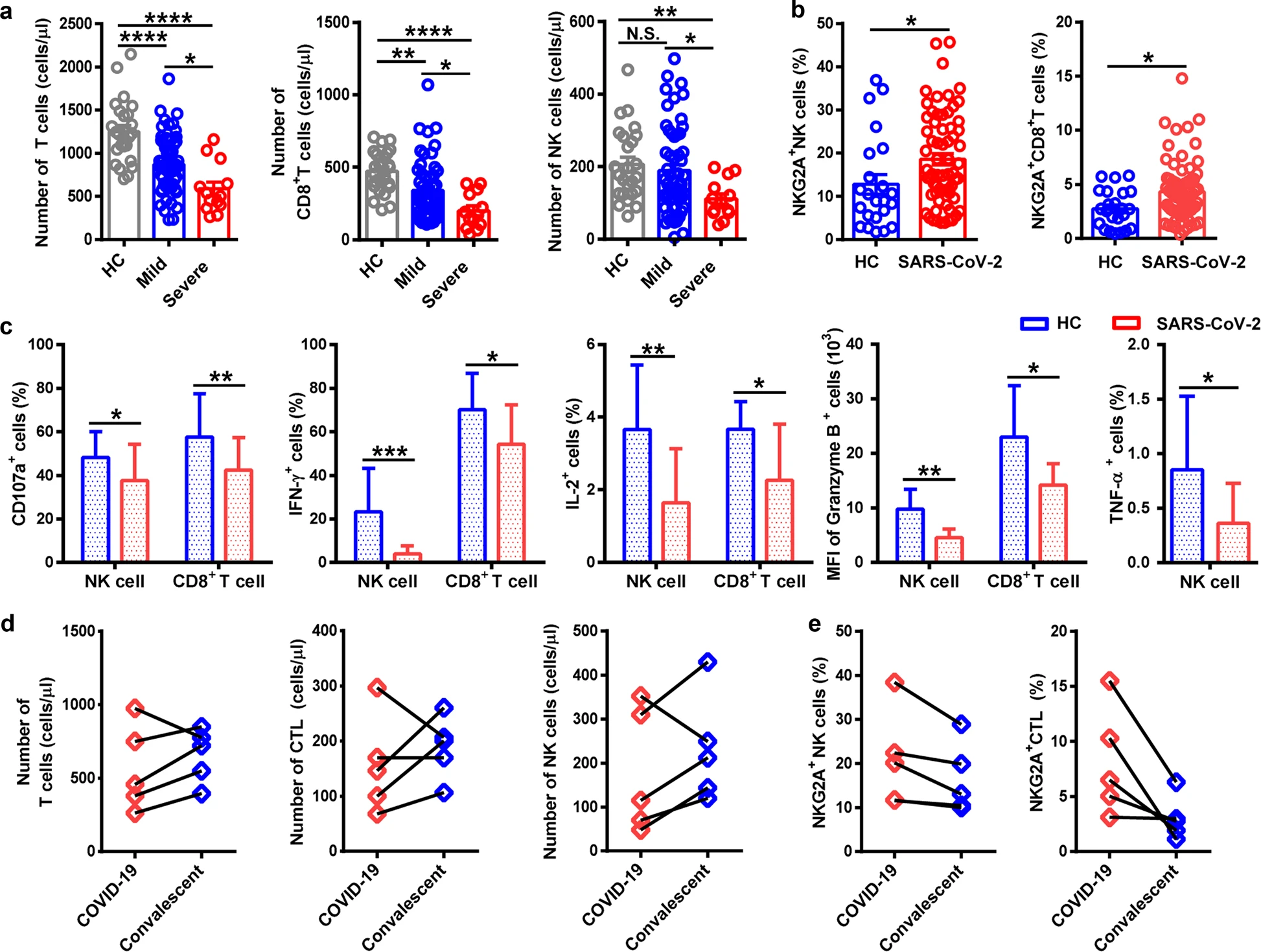
I reported previously on observed functional exhaustion of T cells in patients with severe COVID-19 (see 4/13 updates). In that study, Diao et al. noted increased expression of the exhaustion markers PD-1 and Tim-3 on T cells that increased with the severity of the disease and recovered to normal levels in the convalescent period (Diao et al., 2020). A similar study out of China observed significantly increased expression of another functional exhaustion marker NKG2A on CD8+ T cells as well as on natural killer (NK) cells in severe compared to non-severe cases (Zheng et al., 2020). The measurement of NKG2A was confirmed by findings of decreased intracellular CD107a, IFN-γ, IL-2, and TNF-α in both NK and CD8+ T cells. Additionally, treatment with antiviral therapy (Kaletra™) and interferon, resulted in decreased counts of NKG2A+ NK and cytotoxic T lymphocytes (CTLs) in the convalescent period in all patients observed. The functional exhaustion of immune cells – lymphocytes in particular – indicates a suppression of immune function that may play a large role in the severity of the coronavirus disease. Whether the SARS-CoV-2 virus itself mediates this suppression directly, or whether it is the result of the host inflammatory response remains to be determined. Exhaustion of NK cells in addition to T cells indicates that the immune suppression seen in COVID-19 may be more extensive than is currently understood.
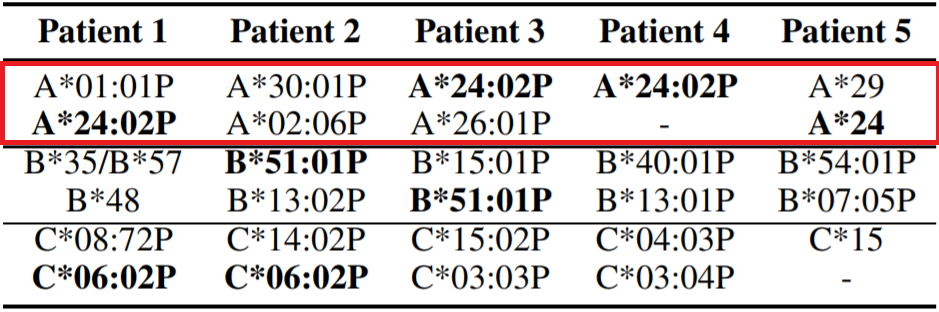
As we look toward the future of COVID-19 prevention, we must take into account the nature of comorbidities and underlying genetic conditions that predispose a certain population to contracting the disease. Human Leukocyte Antigen (HLA) proteins are molecules that play the role of the major histocompatibility complex (MHC) in humans, and function in presenting antigen to the immune system for induction of an immune response (Silvestre, et al., 1970). The HLA gene family is a major contributor to what makes each person unique on a molecular/genetic level, as the polygeny and polymorphism of the class of molecules allows for expression of a unique subset in each person. Because of their direct involvement in the induction of immune responses, specific HLA alleles have been shown to be factors contributing to both susceptibility and resistance to various diseases (Hudson, Allen, 2016).
Using high-throughput proprietary screening software HLAminer™, Warren and Birol retrospectively screened bronchoalveolar lavage fluid of five patients of Chinese ethnicity who had been directly exposed to the seafood market in Wuhan at the epicenter of the outbreak. There was no classification of the severity of disease in any of the five patient samples. They discovered that four of the five patients tested expressed the HLA allele HLA-A*24:02 (Warren, Birol, 2020). The authors note that the prevalence of this specific HLA allele in the general Chinese population is known to be roughly 17.2%, a figure that is statistically significantly different from the observed 80% occurrence in the screened patients. They hypothesize that these data indicate HLA-A*24:02 to be a factor in heightened susceptibility to infection by SARS-CoV-2. Interestingly, this same allele has been described as a risk factor for the development of type I diabetes, which has itself been reported as a risk factor for severe COVID-19 (Noble, et al., 2002; Guo, et al., 2020; Fang, Karakiulakis, Roth, 2020).
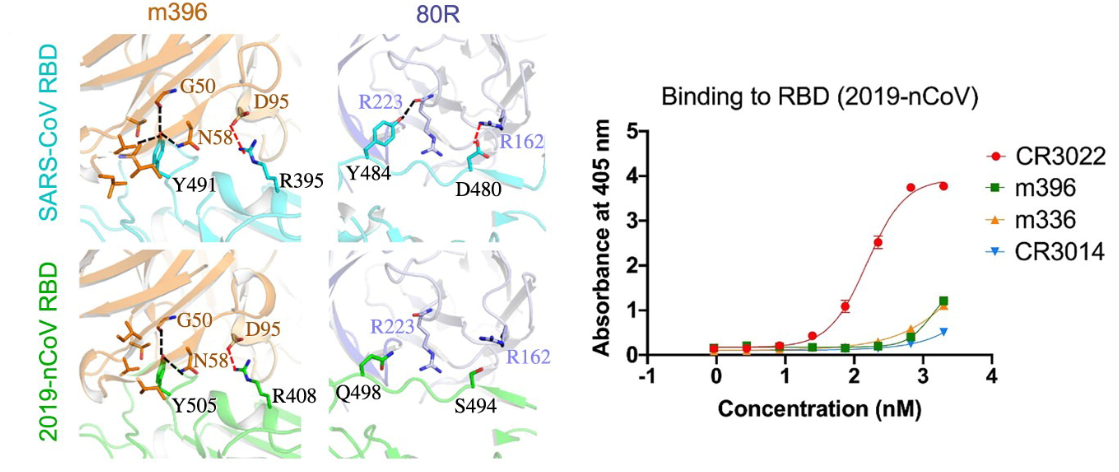
Production of monoclonal antibodies (mAbs) is common as both a laboratory technique and for therapeutic use, largely against tumors (Waldmann, 1991; Weiner, Surana, Wang, 2010). To this end, mAbs developed for use against SARS-CoV and MERS-CoV were tested against the receptor binding domain (RBD) of the surface spike protein of SARS-CoV-2 (Tian, et al., 2020). The goal of the study was to determine potential candidates already in existence for use in the development of a vaccine or antiviral treatment for SARS-CoV-2. Three SARS-CoV and one MERS-CoV mAbs were tested for their affinity to the RBD of SARS-CoV-2 spike protein. It was determined that only one, SARS-CoV mAb CR3022, bound with significant affinity to SARS-CoV-2. When CR3022 was tested concurrently with ACE2 – the putative SARS-CoV-2 target – it demonstrated no competition of binding, indicating that the mAb binds the spike protein at a distinct epitope. Additionally, the monoclonal antibody m396, one of the mAbs with the highest affinity for SARS-CoV, showed no significant binding to SARS-CoV-2, highlighting the genetic and molecular distinction between the two viruses. This was postulated to be due to a difference in residues at several positions along the length of the RBD: slight discrepancies in the positions of an arginine and a tyrosine appear to play a large role in destabilizing the interaction between the RBD and m396 in SARS-CoV-2 (note the arginine at position 395 and tyrosine at position 491 [R395, Y491, respectively] in the SARS-CoV RBD and the arginine at position 408 and tyrosine at position 505 [R408, Y505] in the SARS-CoV-2 RBD in the orange model in the figure below).
Written by: Parker Davis
Edited by: Jina Zhou and Esther Melamed
4/27/2020
References
Hudson, L. E., & Allen, R. L. (2016). Leukocyte Ig-like receptors – A Model for MHC class i disease associations. Frontiers in Immunology, 7(JUL), 281. https://doi.org/10.3389/fimmu.2016.00281
Noble, J. A., Valdes, A. M., Bugawan, T. L., Apple, R. J., Thomson, G., & Erlich, H. A. (2002). The HLA class I A locus affects susceptibility to type 1 diabetes. Human Immunology, 63(8), 657–664. https://doi.org/10.1016/S0198-8859(02)00421-4
Tian, X., Li, C., Huang, A., Xia, S., Lu, S., Shi, Z., Lu, L., Jiang, S., Yang, Z., Wu, Y., & Ying, T. (2020). Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. In Emerging Microbes and Infections (Vol. 9, Issue 1, pp. 382–385). Taylor and Francis Ltd. https://doi.org/10.1080/22221751.2020.1729069
Waldmann, T. A. (1991). Monoclonal antibodies in diagnosis and therapy. Science, 252(5013), 1657–1662. https://doi.org/10.1126/science.2047874
Warren, R. L., & Birol, I. (2020). HLA predictions from the bronchoalveolar lavage fluid samples of five patients at the early stage of the Wuhan seafood market COVID-19 outbreak. http://arxiv.org/abs/2004.07108
Weiner, L. M., Surana, R., & Wang, S. (2010). Monoclonal antibodies: Versatile platforms for cancer immunotherapy. In Nature Reviews Immunology (Vol. 10, Issue 5, pp. 317–327). Nature Publishing Group. https://doi.org/10.1038/nri2744
Zheng, M., Gao, Y., Wang, G., Song, G., Liu, S., Sun, D., Xu, Y., & Tian, Z. (2020). Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cellular & Molecular Immunology, 1–3. https://doi.org/10.1038/s41423-020-0402-2
Leave a Reply