Study of Poly(ADP-ribose) Polymerase-1
Poly(ADP-ribose) polymerase-1 (PARP-1) is an ubiquitous human enzyme that catalyzes the synthesis of the branched polymer poly(ADP-ribose) using NAD+ as the building block. This posttranslational modification typically occurs on the glutamate residues of the target proteins. Many protein substrates, including histones, transcription factors, DNA repair proteins, and PARP-1 itself have been identified. Because poly(ADP-ribosyl)ation significantly alters the DNA binding properties of the target proteins, PARP-1 plays a pivotal role in both DNA repair and the regulation of gene transcription. PARP-1 activity is elaborately regulated in the cell by many factors, such as DNA damage, caspases, and the intracellular NAD+ concentration. For example, inactivation of PARP-1 by caspase-3 cleavage is one of the key events of apoptosis, while over-activation of PARP-1 leads to ATP-depletion and subsequently to necrosis. The goals of this project are two-fold: to study how PARP-1 activity is regulated and to gain mechanistic insight useful for designing specific inhibitors for PARP-1.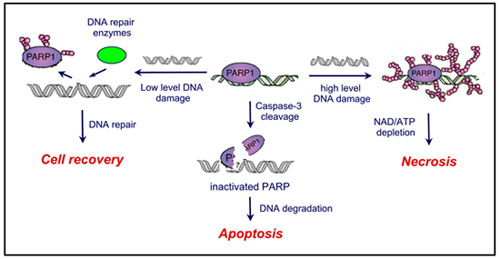
To elucidate the mechanism of PARP-1 activation, we decided to examine the function of different PARP-1 domains. As illustrated below, PARP-1 has been dissected into six smaller regions, named A through F, based on sequence homology with previously identified functional modules present in other enzymes. To study the potential role of each domain of PARP-1 in self-regulation, we have constructed a set of PARP-1 domain deletion mutants. In vitro activity assays, protein-protein interaction studies, and analysis of poly(ADP-ribosyl)ation of these mutants have been carried out to investigate the mechanism of PARP-1 activation. Our results indicate that:
- Domain A is required for DNA-dependent activation through zinc finger recognition of DNA-breaks;
- Domain C plays a important role in PARP-1 activation via an allosteric mechanism;
- PARP-1 is self-modified at a number of glutamate residues present throughout the PARP-1 structure.
Detailed studies to identify allosteric sites on PARP-1, which regulate its activity through domain-domain interactions, are ongoing. Disruption of these interactions may cause inactivation of PARP-1 and provide a new strategy for the design of specific PARP-1 inhibitors.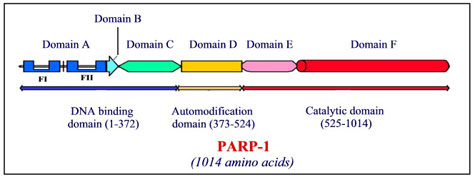
We are also studying PARP-1 trans-modification and interaction of PARP-1 with other proteins. The specific PARP-1/protein interactions under investigation include those between PARP-1/AIF and PARP-1/NMNAT. A high-throughput method to identify PARP-1 protein substrates has been developed to probe the biological functions of PARP-1 in vivo. In addition, we have synthesized a series of fluorinated NAD+ analogues to study the mechanism of the polymerization reaction and the structure-activity relationship of the poly(ADP-ribose) polymers. Several of these analogues can be used as specific inhibitors to stop either the elongation or the branching reactions.

